Gene
KWMTBOMO12009 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004501
Annotation
transcription_elongation_factor_B_polypeptide_1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.541
Sequence
CDS
ATGTCCGAGCAACCTTCTGTAACTAATGTTGCTGATGAACAACATTCAGCTAGTGGATCTGGCAGTATCGGTGGTGAAGAGAAGGTGTATGGAGGATGTGAAGGTCCAGATGCTATGTATGTGAAACTGGTATCCTCTGATGGACATGAGTTCATTGTGAAGAGGGAGCATGCTCTTATATCAGGCACTATTAAGGCTATGCTCAGCGGACCGGGCCAGTTTGCTGAAAATGAAGCTAACGAGGTCAACTTTAGAGAGATACCATCACACGTACTCCAAAAAGTCTGCATGTACTTCACGTACAAGGTACGCTATACCAACTCGTCTACAGAGATACCAGAGTTTCCGATCGCTCCCGAGATCGCCCTCGAAGTGTTGATGGCTGCCAACTTCCTCGACTGTTAA
Protein
MSEQPSVTNVADEQHSASGSGSIGGEEKVYGGCEGPDAMYVKLVSSDGHEFIVKREHALISGTIKAMLSGPGQFAENEANEVNFREIPSHVLQKVCMYFTYKVRYTNSSTEIPEFPIAPEIALEVLMAANFLDC
Summary
Similarity
Belongs to the SKP1 family.
Uniprot
Q1HQ04
S4P807
A0A212FAC6
I4DMV3
A0A194QQR5
I4DK57
+ More
A0A0L7LVS3 A0A194QGT1 A0A1Y1KWR8 V5H0Q4 A0A2P8XVD1 A0A2H1VDA5 D6WQT6 T1JI02 A0A2M3ZB20 A0A1E1WDJ1 A0A2A4JI60 A0A2W1BI45 A0A3R7NDT7 A0A210QBT3 A0A182MV84 A0A0P4WT88 T1DPS6 A0A2M4C3U6 A0A182FLN0 A0A1Q3FUE4 A0A084VRL6 N6SWB9 A0A194AK23 A0A1S3INC6 A0A1W4WVL9 A0A182XC27 A0A182RQZ8 A0A182QZC6 A0A182V6I4 A0A182J5R8 A0A182Y5X5 A0A182K615 A0A182LP82 A0A182PU87 A0A182W2F9 A0A182U6W0 A0A182HNC5 A0A182LS43 Q7PSG2 Q17C74 A0A023EEX0 A0A0E9X2W9 Q502M9 A0A2K5PML1 A0A0K8TP85 A0A2J7QHF8 F7D5C0 A0A286YBA4 B4KME6 J3JVJ3 W5JWE1 A0A3S2PR98 A0A182WGQ4 A0A0M3QVN0 S4RTH7 A0A093G207 A0A091QPE2 A0A091DU57 A0A2P4SSA5 F7C1R4 U3IAW3 A0A0N8K118 H0ZMU1 H0XY64 B4LNW7 A0A3B0JPV5 B5DUR2 B4GIM9 B4J624 A0A2Y9F7P6 G1NG91 A0A151MN60 A0A226NLZ2 A0A3L8T0S4 C1BIJ8 A0A218UPE1 A0A3M0KQ91 K7F7A1 A0A0B8RTH2 A0A0Q3X6W3 A0A0E9WT96 A0A2I0MWE7 A0A1W7RCU1 T1D843 C1C460 J3S8L6 Q5ZJT7 A0A0P7Z789 C1BEZ0 Q6PHW7 A0A1U7RJA2 Q28H26 U3ETF7 B9EN56
A0A0L7LVS3 A0A194QGT1 A0A1Y1KWR8 V5H0Q4 A0A2P8XVD1 A0A2H1VDA5 D6WQT6 T1JI02 A0A2M3ZB20 A0A1E1WDJ1 A0A2A4JI60 A0A2W1BI45 A0A3R7NDT7 A0A210QBT3 A0A182MV84 A0A0P4WT88 T1DPS6 A0A2M4C3U6 A0A182FLN0 A0A1Q3FUE4 A0A084VRL6 N6SWB9 A0A194AK23 A0A1S3INC6 A0A1W4WVL9 A0A182XC27 A0A182RQZ8 A0A182QZC6 A0A182V6I4 A0A182J5R8 A0A182Y5X5 A0A182K615 A0A182LP82 A0A182PU87 A0A182W2F9 A0A182U6W0 A0A182HNC5 A0A182LS43 Q7PSG2 Q17C74 A0A023EEX0 A0A0E9X2W9 Q502M9 A0A2K5PML1 A0A0K8TP85 A0A2J7QHF8 F7D5C0 A0A286YBA4 B4KME6 J3JVJ3 W5JWE1 A0A3S2PR98 A0A182WGQ4 A0A0M3QVN0 S4RTH7 A0A093G207 A0A091QPE2 A0A091DU57 A0A2P4SSA5 F7C1R4 U3IAW3 A0A0N8K118 H0ZMU1 H0XY64 B4LNW7 A0A3B0JPV5 B5DUR2 B4GIM9 B4J624 A0A2Y9F7P6 G1NG91 A0A151MN60 A0A226NLZ2 A0A3L8T0S4 C1BIJ8 A0A218UPE1 A0A3M0KQ91 K7F7A1 A0A0B8RTH2 A0A0Q3X6W3 A0A0E9WT96 A0A2I0MWE7 A0A1W7RCU1 T1D843 C1C460 J3S8L6 Q5ZJT7 A0A0P7Z789 C1BEZ0 Q6PHW7 A0A1U7RJA2 Q28H26 U3ETF7 B9EN56
Pubmed
19121390
23622113
22118469
22651552
26354079
26227816
+ More
28004739 29403074 18362917 19820115 28756777 28812685 24438588 23537049 25244985 20966253 12364791 17510324 24945155 26483478 25613341 23594743 26369729 19892987 17994087 18057021 22516182 20920257 23761445 20360741 15632085 20838655 22293439 30282656 17381049 25476704 23371554 26358130 23758969 23025625 15592404 15642098 27762356 20431018 23915248 20433749
28004739 29403074 18362917 19820115 28756777 28812685 24438588 23537049 25244985 20966253 12364791 17510324 24945155 26483478 25613341 23594743 26369729 19892987 17994087 18057021 22516182 20920257 23761445 20360741 15632085 20838655 22293439 30282656 17381049 25476704 23371554 26358130 23758969 23025625 15592404 15642098 27762356 20431018 23915248 20433749
EMBL
BABH01022140
DQ443248
ABF51337.1
GAIX01004359
JAA88201.1
AGBW02009503
+ More
OWR50673.1 AK402621 BAM19243.1 KQ461181 KPJ07689.1 AK401675 BAM18297.1 JTDY01000033 KOB79286.1 KQ459053 KPJ04145.1 GEZM01075351 JAV64125.1 GALX01000619 JAB67847.1 PYGN01001292 PYGN01000792 PSN35946.1 PSN40759.1 ODYU01001925 SOQ38807.1 KQ971354 EFA07009.1 JH431967 GGFM01004951 MBW25702.1 GDQN01008849 GDQN01008794 GDQN01006047 JAT82205.1 JAT82260.1 JAT85007.1 NWSH01001376 PCG71479.1 KZ150043 PZC74528.1 QCYY01000534 ROT84479.1 NEDP02004236 OWF46193.1 AXCM01003888 GDRN01021211 JAI67809.1 GAMD01003089 JAA98501.1 GGFJ01010859 MBW60000.1 GFDL01003887 JAV31158.1 ATLV01015746 KE525033 KFB40610.1 APGK01054165 KB741247 KB632281 ENN72054.1 ERL91381.1 GELH01000604 GELH01000603 JAS03669.1 AXCN02002036 APCN01001904 AXCM01006181 AXCM01019587 AAAB01008834 EAA05684.3 CH477311 EAT43917.1 JXUM01056140 GAPW01006197 KQ561898 JAC07401.1 KXJ77225.1 GBXM01011490 JAH97087.1 CR788254 BC095636 BC165485 AAH95636.1 AAI65485.1 GDAI01001444 JAI16159.1 NEVH01013982 PNF27983.1 CH933808 EDW09834.1 KRG04929.1 BT127261 AEE62223.1 ADMH02000225 ETN67309.1 CM012456 RVE58854.1 CP012524 ALC42689.1 KL215087 KFV63163.1 KK801951 KFQ29400.1 KN122025 KFO34028.1 PPHD01025902 POI26993.1 JARO02002046 KPP73697.1 ABQF01005430 ABQF01005431 ABQF01005432 AAQR03158414 AAQR03158415 AAQR03158416 AAQR03158417 AAQR03158418 CH940648 EDW61136.1 OUUW01000001 SPP75679.1 CH674335 EDY71631.1 CH479183 EDW36349.1 CH916367 EDW01882.1 AKHW03005657 KYO25964.1 MCFN01000009 OXB68725.1 QUSF01000001 RLW13238.1 BT074427 ACO08851.1 MUZQ01000198 OWK55526.1 QRBI01000104 RMC15273.1 AGCU01142303 AGCU01142304 AGCU01142305 GBSH01002706 JAG66321.1 LMAW01000392 KQL59191.1 GBXM01015864 JAH92713.1 AKCR02000001 PKK34006.1 GDAY02002512 JAV48955.1 GAAZ01002499 JAA95444.1 BT081639 ACO51770.1 JU174432 GBEX01003399 AFJ49958.1 JAI11161.1 AADN05000031 AJ720347 CAG32006.1 JARO02001072 KPP76565.1 BT073169 BT073552 ACO07593.1 BC055268 CM004477 CM004476 AAH55268.1 OCT74816.1 OCT76632.1 AAMC01065653 BC135463 CR761092 AAI35464.1 CAJ81641.1 GAEP01000503 GBEW01001025 JAB54318.1 JAI09340.1 BT057081 ACM08953.1
OWR50673.1 AK402621 BAM19243.1 KQ461181 KPJ07689.1 AK401675 BAM18297.1 JTDY01000033 KOB79286.1 KQ459053 KPJ04145.1 GEZM01075351 JAV64125.1 GALX01000619 JAB67847.1 PYGN01001292 PYGN01000792 PSN35946.1 PSN40759.1 ODYU01001925 SOQ38807.1 KQ971354 EFA07009.1 JH431967 GGFM01004951 MBW25702.1 GDQN01008849 GDQN01008794 GDQN01006047 JAT82205.1 JAT82260.1 JAT85007.1 NWSH01001376 PCG71479.1 KZ150043 PZC74528.1 QCYY01000534 ROT84479.1 NEDP02004236 OWF46193.1 AXCM01003888 GDRN01021211 JAI67809.1 GAMD01003089 JAA98501.1 GGFJ01010859 MBW60000.1 GFDL01003887 JAV31158.1 ATLV01015746 KE525033 KFB40610.1 APGK01054165 KB741247 KB632281 ENN72054.1 ERL91381.1 GELH01000604 GELH01000603 JAS03669.1 AXCN02002036 APCN01001904 AXCM01006181 AXCM01019587 AAAB01008834 EAA05684.3 CH477311 EAT43917.1 JXUM01056140 GAPW01006197 KQ561898 JAC07401.1 KXJ77225.1 GBXM01011490 JAH97087.1 CR788254 BC095636 BC165485 AAH95636.1 AAI65485.1 GDAI01001444 JAI16159.1 NEVH01013982 PNF27983.1 CH933808 EDW09834.1 KRG04929.1 BT127261 AEE62223.1 ADMH02000225 ETN67309.1 CM012456 RVE58854.1 CP012524 ALC42689.1 KL215087 KFV63163.1 KK801951 KFQ29400.1 KN122025 KFO34028.1 PPHD01025902 POI26993.1 JARO02002046 KPP73697.1 ABQF01005430 ABQF01005431 ABQF01005432 AAQR03158414 AAQR03158415 AAQR03158416 AAQR03158417 AAQR03158418 CH940648 EDW61136.1 OUUW01000001 SPP75679.1 CH674335 EDY71631.1 CH479183 EDW36349.1 CH916367 EDW01882.1 AKHW03005657 KYO25964.1 MCFN01000009 OXB68725.1 QUSF01000001 RLW13238.1 BT074427 ACO08851.1 MUZQ01000198 OWK55526.1 QRBI01000104 RMC15273.1 AGCU01142303 AGCU01142304 AGCU01142305 GBSH01002706 JAG66321.1 LMAW01000392 KQL59191.1 GBXM01015864 JAH92713.1 AKCR02000001 PKK34006.1 GDAY02002512 JAV48955.1 GAAZ01002499 JAA95444.1 BT081639 ACO51770.1 JU174432 GBEX01003399 AFJ49958.1 JAI11161.1 AADN05000031 AJ720347 CAG32006.1 JARO02001072 KPP76565.1 BT073169 BT073552 ACO07593.1 BC055268 CM004477 CM004476 AAH55268.1 OCT74816.1 OCT76632.1 AAMC01065653 BC135463 CR761092 AAI35464.1 CAJ81641.1 GAEP01000503 GBEW01001025 JAB54318.1 JAI09340.1 BT057081 ACM08953.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000037510
UP000053268
UP000245037
+ More
UP000007266 UP000218220 UP000283509 UP000242188 UP000075883 UP000069272 UP000030765 UP000019118 UP000030742 UP000085678 UP000192223 UP000076407 UP000075900 UP000075886 UP000075903 UP000075880 UP000076408 UP000075881 UP000075882 UP000075885 UP000075920 UP000075902 UP000075840 UP000007062 UP000008820 UP000069940 UP000249989 UP000000437 UP000233040 UP000235965 UP000002281 UP000009192 UP000000673 UP000092553 UP000245300 UP000053875 UP000028990 UP000008225 UP000034805 UP000007754 UP000005225 UP000008792 UP000268350 UP000001819 UP000008744 UP000001070 UP000001645 UP000050525 UP000198323 UP000276834 UP000197619 UP000269221 UP000007267 UP000051836 UP000053872 UP000000539 UP000192224 UP000186698 UP000189705 UP000008143 UP000087266
UP000007266 UP000218220 UP000283509 UP000242188 UP000075883 UP000069272 UP000030765 UP000019118 UP000030742 UP000085678 UP000192223 UP000076407 UP000075900 UP000075886 UP000075903 UP000075880 UP000076408 UP000075881 UP000075882 UP000075885 UP000075920 UP000075902 UP000075840 UP000007062 UP000008820 UP000069940 UP000249989 UP000000437 UP000233040 UP000235965 UP000002281 UP000009192 UP000000673 UP000092553 UP000245300 UP000053875 UP000028990 UP000008225 UP000034805 UP000007754 UP000005225 UP000008792 UP000268350 UP000001819 UP000008744 UP000001070 UP000001645 UP000050525 UP000198323 UP000276834 UP000197619 UP000269221 UP000007267 UP000051836 UP000053872 UP000000539 UP000192224 UP000186698 UP000189705 UP000008143 UP000087266
PRIDE
Pfam
PF03931 Skp1_POZ
Interpro
SUPFAM
SSF54695
SSF54695
ProteinModelPortal
Q1HQ04
S4P807
A0A212FAC6
I4DMV3
A0A194QQR5
I4DK57
+ More
A0A0L7LVS3 A0A194QGT1 A0A1Y1KWR8 V5H0Q4 A0A2P8XVD1 A0A2H1VDA5 D6WQT6 T1JI02 A0A2M3ZB20 A0A1E1WDJ1 A0A2A4JI60 A0A2W1BI45 A0A3R7NDT7 A0A210QBT3 A0A182MV84 A0A0P4WT88 T1DPS6 A0A2M4C3U6 A0A182FLN0 A0A1Q3FUE4 A0A084VRL6 N6SWB9 A0A194AK23 A0A1S3INC6 A0A1W4WVL9 A0A182XC27 A0A182RQZ8 A0A182QZC6 A0A182V6I4 A0A182J5R8 A0A182Y5X5 A0A182K615 A0A182LP82 A0A182PU87 A0A182W2F9 A0A182U6W0 A0A182HNC5 A0A182LS43 Q7PSG2 Q17C74 A0A023EEX0 A0A0E9X2W9 Q502M9 A0A2K5PML1 A0A0K8TP85 A0A2J7QHF8 F7D5C0 A0A286YBA4 B4KME6 J3JVJ3 W5JWE1 A0A3S2PR98 A0A182WGQ4 A0A0M3QVN0 S4RTH7 A0A093G207 A0A091QPE2 A0A091DU57 A0A2P4SSA5 F7C1R4 U3IAW3 A0A0N8K118 H0ZMU1 H0XY64 B4LNW7 A0A3B0JPV5 B5DUR2 B4GIM9 B4J624 A0A2Y9F7P6 G1NG91 A0A151MN60 A0A226NLZ2 A0A3L8T0S4 C1BIJ8 A0A218UPE1 A0A3M0KQ91 K7F7A1 A0A0B8RTH2 A0A0Q3X6W3 A0A0E9WT96 A0A2I0MWE7 A0A1W7RCU1 T1D843 C1C460 J3S8L6 Q5ZJT7 A0A0P7Z789 C1BEZ0 Q6PHW7 A0A1U7RJA2 Q28H26 U3ETF7 B9EN56
A0A0L7LVS3 A0A194QGT1 A0A1Y1KWR8 V5H0Q4 A0A2P8XVD1 A0A2H1VDA5 D6WQT6 T1JI02 A0A2M3ZB20 A0A1E1WDJ1 A0A2A4JI60 A0A2W1BI45 A0A3R7NDT7 A0A210QBT3 A0A182MV84 A0A0P4WT88 T1DPS6 A0A2M4C3U6 A0A182FLN0 A0A1Q3FUE4 A0A084VRL6 N6SWB9 A0A194AK23 A0A1S3INC6 A0A1W4WVL9 A0A182XC27 A0A182RQZ8 A0A182QZC6 A0A182V6I4 A0A182J5R8 A0A182Y5X5 A0A182K615 A0A182LP82 A0A182PU87 A0A182W2F9 A0A182U6W0 A0A182HNC5 A0A182LS43 Q7PSG2 Q17C74 A0A023EEX0 A0A0E9X2W9 Q502M9 A0A2K5PML1 A0A0K8TP85 A0A2J7QHF8 F7D5C0 A0A286YBA4 B4KME6 J3JVJ3 W5JWE1 A0A3S2PR98 A0A182WGQ4 A0A0M3QVN0 S4RTH7 A0A093G207 A0A091QPE2 A0A091DU57 A0A2P4SSA5 F7C1R4 U3IAW3 A0A0N8K118 H0ZMU1 H0XY64 B4LNW7 A0A3B0JPV5 B5DUR2 B4GIM9 B4J624 A0A2Y9F7P6 G1NG91 A0A151MN60 A0A226NLZ2 A0A3L8T0S4 C1BIJ8 A0A218UPE1 A0A3M0KQ91 K7F7A1 A0A0B8RTH2 A0A0Q3X6W3 A0A0E9WT96 A0A2I0MWE7 A0A1W7RCU1 T1D843 C1C460 J3S8L6 Q5ZJT7 A0A0P7Z789 C1BEZ0 Q6PHW7 A0A1U7RJA2 Q28H26 U3ETF7 B9EN56
PDB
1VCB
E-value=2.90538e-55,
Score=538
Ontologies
GO
PANTHER
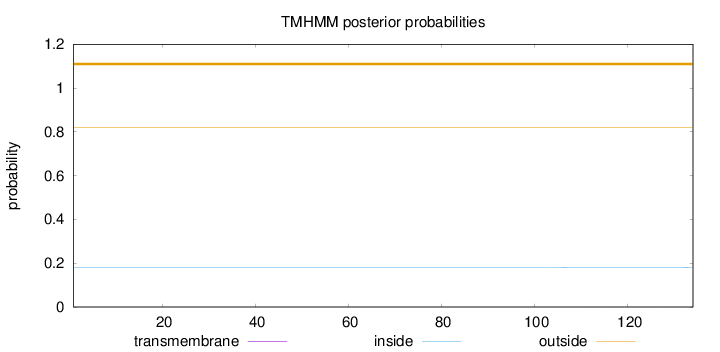
Topology
Length:
134
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00848
Exp number, first 60 AAs:
0.00014
Total prob of N-in:
0.17988
outside
1 - 134
Population Genetic Test Statistics
Pi
71.810661
Theta
125.168028
Tajima's D
-1.0089
CLR
177.430601
CSRT
0.138793060346983
Interpretation
Uncertain
