Gene
KWMTBOMO11991
Pre Gene Modal
BGIBMGA004373
Annotation
PREDICTED:_N6-adenosine-methyltransferase_70_kDa_subunit_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.459
Sequence
CDS
ATGTCGGACGCTTGGGAGGAAATACAAGCTGTAAAAAGCAAAAGGAAAAGTTTCCGTGAAAAGTTGGAGAAACGAAAAAAGGAACGCCAAAGTATCTTGAGCTCTAGTTTGGTCAGTGGCGAGAAATCTGATGCCTCTCCTAATAAAGATCGATCACTTGGTAGCCCGGCTTCAAAATCAGAAGCTTCAACTTTGCAAGAAAAGTCAAAACCAATCGTTCCACCAGCATCATTGGAAGTGCGTCTATTACAAGTTCTATCAGATATGCAATTGCAGTTACCTGCTACTGCCAGTGCTCTGCTACCAGGACTTGGAGATGTGGACACGGAGATTGTAGCAAACCTGTTACAGAAGTTTGCAACACAGAAGTTGATCACTATAAGAGAAAAAACTGAAGTAACAACAGATGCTGACGTTGAGGTAGTGAGTGCAGAGTCAGGGAGGCTTGCGGCCGTTCTTGCAGCTCTGATGGAAGAAGAAACCACGACAACTAGCAACAAAAGAAAAGGTGAGAGTCCCATTGAAGATACATCAATTACAAAAGCTCCTAAGATGTCCACCGAGGAAAAGAAGTCTGGCAAGAGCGACACAGATATTATGTCCCTCCTAGCGATGCCTTCAAGTAGAGAGAAAGCAGTGAAGAGAGTCGGTGAGGAAATAATGGATTTATTGAGCAAGCCCACAGCTAAAGAGAGGTCCTTAGCTGACAAATTCAAAAGCCAAGGAGGTGCTCAAGTAATGGAGTTCTGTCCGCACGGTACCAGAGCGGAGTGCGGTAGAGCTATCAGCCCTAACAGTGCACAAGACGGTACCACCAACACCTGCAAAAAGTTACATTTCAAAAAGATCATACAGGGTCACACCGACGAAACCTTGGGTGACTGTTCATTCCTGAACACGTGCTTTCATATGGACAGCTGCAAGTACGTTCATTACGAAGTCGACAACACCGATCCAAATGCGAACACTAAGTCAGTGGAGGCGCTGACGAAGTCTGTGCAAAATGGTACAAGTTCGATATCGAAGTGCTTGAATACAGACGGTGTTTTGACTCTGACGCCTCCGCAGTGGATACAGTGCGACCTTAGATATTTGGACATGACGTTCTTAGGTAAATTCGCCGTGATAATGGCTGACCCGCCTTGGGATATCCATATGGAGCTGCCCTATGGCACTATGTCGGACGACGAGATGCGGTGCCTCGGAATACCGCAACTGCAGGACAGCGGCCTCATATTCCTGTGGGTCACGGGCAGAGCGATGGAATTGGGTCGCGAATGCCTCAAGCTGTGGGGCTACGAGCGCGTCGACGAGCTGATATGGGTGAAGACGAATCAACTGCAACGCATCATCAGAACCGGCCGGACCGGGCACTGGCTGAACCACGGGAAAGAACATTGTCTTGTTGGCATGAAAGGCAGCCCTGAGAATTTGAATCGGGGTCTGGACTGTGACGTCATCGTAGCCGAAGTTAGGGCGACTTCACACAAACCCGACGAGATATACGGCATTATCGAGAGGCTCAGTCCGGGTACAAGGAAGATCGAATTATTCGGTCGCCCACATAACGTTCAACCGAATTGGATAACTCTGGGGAACCAGGTGGACGGCGTGAACCTTGTGGATCCGGACCTGATAGTCGCCTTCAAGAAACGTTATCCGGACGGCAACTGCATGGCGCCGCCGCCGCCAGACCCGGGACTCACTTAG
Protein
MSDAWEEIQAVKSKRKSFREKLEKRKKERQSILSSSLVSGEKSDASPNKDRSLGSPASKSEASTLQEKSKPIVPPASLEVRLLQVLSDMQLQLPATASALLPGLGDVDTEIVANLLQKFATQKLITIREKTEVTTDADVEVVSAESGRLAAVLAALMEEETTTTSNKRKGESPIEDTSITKAPKMSTEEKKSGKSDTDIMSLLAMPSSREKAVKRVGEEIMDLLSKPTAKERSLADKFKSQGGAQVMEFCPHGTRAECGRAISPNSAQDGTTNTCKKLHFKKIIQGHTDETLGDCSFLNTCFHMDSCKYVHYEVDNTDPNANTKSVEALTKSVQNGTSSISKCLNTDGVLTLTPPQWIQCDLRYLDMTFLGKFAVIMADPPWDIHMELPYGTMSDDEMRCLGIPQLQDSGLIFLWVTGRAMELGRECLKLWGYERVDELIWVKTNQLQRIIRTGRTGHWLNHGKEHCLVGMKGSPENLNRGLDCDVIVAEVRATSHKPDEIYGIIERLSPGTRKIELFGRPHNVQPNWITLGNQVDGVNLVDPDLIVAFKKRYPDGNCMAPPPPDPGLT
Summary
Similarity
Belongs to the MT-A70-like family.
Uniprot
H9J4D4
A0A2A4KAR4
A0A194QFA7
A0A1B6JWU7
E0VST8
A0A1Y1NB44
+ More
A0A2J7PSI8 A0A067R9A5 A0A1B0DCU0 A0A1W4X4V1 A0A2P8Y580 A0A1B6DGV4 D6WRZ1 A0A087UQB0 A0A336K036 A0A2P6KPA9 A0A0N0BDR7 B4HGG4 A0A232EUI7 E2C5I8 A0A0L7RBI6 A0A154PGK9 A0A0J7KQ31 K7IWZ1 A0A1E1X5B7 A0A026WCD5 A0A087ZTT3 E9IGP1 A0A0P4VIY7 A0A2A3EBZ7 T1J262 A0A195BR29 A0A158NRS6 A0A0V0GBW6 E2A9M3 A0A195D150 A0A023F3T6 A0A195EG26 A0A151X3D4 A0A2S2NG63 A0A1J1ISQ6 L7M7H1 F4WQT2 A0A131YN03 A0A195ERQ2 A0A2S2QW06 A0A224YVY9 A0A0P6I1H8 A0A023GFK5 A0A0T6B077 A0A0N8DCV0 A0A131XFC5 A0A0P5B504 A0A1V9XBW1 A0A1B3PDH6 J9JK00 A0A0P5WTI7 E9FZ30 A0A0P5AFU4 A0A146KWJ1 A0A0N8A345 A0A0P5N6G7 A0A0A9Y4S1 A0A131Y1C5 A0A147BCX0 A0A1B3PDC1 A0A0K8TB88 A0A0N8CEV7 A0A0P5C603 A0A0P5C5P2 A0A0P5SFJ9 A0A3S3PLS7 A0A0L8G1B4 A0A210PFF4 A0A3S3NJR2 A0A293LPZ7 A0A1Z5KWW9 A0A3S1BYF3 V3ZQ95 A0A0B6ZWL5 A0A0P5W3U3 A0A1S3J0D7 R7T3J2 A0A0P5QIF2 C3YFD7 A0A1L1RJR6 A0A0N8CZC7 K1R9D4 A0A0P5WKY6 A0A1U7S5E4 A0A0P6BT66
A0A2J7PSI8 A0A067R9A5 A0A1B0DCU0 A0A1W4X4V1 A0A2P8Y580 A0A1B6DGV4 D6WRZ1 A0A087UQB0 A0A336K036 A0A2P6KPA9 A0A0N0BDR7 B4HGG4 A0A232EUI7 E2C5I8 A0A0L7RBI6 A0A154PGK9 A0A0J7KQ31 K7IWZ1 A0A1E1X5B7 A0A026WCD5 A0A087ZTT3 E9IGP1 A0A0P4VIY7 A0A2A3EBZ7 T1J262 A0A195BR29 A0A158NRS6 A0A0V0GBW6 E2A9M3 A0A195D150 A0A023F3T6 A0A195EG26 A0A151X3D4 A0A2S2NG63 A0A1J1ISQ6 L7M7H1 F4WQT2 A0A131YN03 A0A195ERQ2 A0A2S2QW06 A0A224YVY9 A0A0P6I1H8 A0A023GFK5 A0A0T6B077 A0A0N8DCV0 A0A131XFC5 A0A0P5B504 A0A1V9XBW1 A0A1B3PDH6 J9JK00 A0A0P5WTI7 E9FZ30 A0A0P5AFU4 A0A146KWJ1 A0A0N8A345 A0A0P5N6G7 A0A0A9Y4S1 A0A131Y1C5 A0A147BCX0 A0A1B3PDC1 A0A0K8TB88 A0A0N8CEV7 A0A0P5C603 A0A0P5C5P2 A0A0P5SFJ9 A0A3S3PLS7 A0A0L8G1B4 A0A210PFF4 A0A3S3NJR2 A0A293LPZ7 A0A1Z5KWW9 A0A3S1BYF3 V3ZQ95 A0A0B6ZWL5 A0A0P5W3U3 A0A1S3J0D7 R7T3J2 A0A0P5QIF2 C3YFD7 A0A1L1RJR6 A0A0N8CZC7 K1R9D4 A0A0P5WKY6 A0A1U7S5E4 A0A0P6BT66
Pubmed
19121390
26354079
20566863
28004739
24845553
29403074
+ More
18362917 19820115 17994087 28648823 20798317 20075255 28503490 24508170 30249741 21282665 27129103 21347285 25474469 25576852 21719571 26830274 28797301 28049606 28327890 21292972 26823975 25401762 29652888 28812685 28528879 23254933 18563158 15592404 22992520
18362917 19820115 17994087 28648823 20798317 20075255 28503490 24508170 30249741 21282665 27129103 21347285 25474469 25576852 21719571 26830274 28797301 28049606 28327890 21292972 26823975 25401762 29652888 28812685 28528879 23254933 18563158 15592404 22992520
EMBL
BABH01022092
BABH01022093
BABH01022094
BABH01022095
BABH01022096
BABH01022097
+ More
NWSH01000004 PCG81004.1 KQ459053 KPJ04167.1 GECU01004019 JAT03688.1 DS235756 EEB16444.1 GEZM01011792 JAV93436.1 NEVH01021928 PNF19301.1 KK852611 KDR20261.1 AJVK01014224 PYGN01000912 PSN39413.1 GEDC01012453 JAS24845.1 KQ971351 EFA06415.1 KK121013 KFM79549.1 UFQS01000041 UFQT01000041 SSW98306.1 SSX18692.1 MWRG01007578 PRD28170.1 KQ435851 KOX70831.1 CH480815 EDW43417.1 NNAY01002117 OXU22029.1 GL452770 EFN76801.1 KQ414617 KOC68203.1 KQ434899 KZC10952.1 LBMM01004397 KMQ92462.1 AAZX01012980 GFAC01004743 JAT94445.1 KK107274 QOIP01000008 EZA53732.1 RLU19300.1 GL763054 EFZ20284.1 GDKW01002553 JAI54042.1 KZ288291 PBC29210.1 JH431796 KQ976424 KYM88593.1 ADTU01024289 GECL01001245 JAP04879.1 GL437918 EFN69825.1 KQ977041 KYN06129.1 GBBI01002627 JAC16085.1 KQ978983 KYN26827.1 KQ982562 KYQ54925.1 GGMR01003574 MBY16193.1 CVRI01000059 CRL03231.1 GACK01005034 JAA60000.1 GL888275 EGI63445.1 GEDV01007903 JAP80654.1 KQ981993 KYN30903.1 GGMS01012712 MBY81915.1 GFPF01009949 MAA21095.1 GDIQ01010969 JAN83768.1 GBBM01003670 JAC31748.1 LJIG01016351 KRT80863.1 GDIP01046247 LRGB01001005 JAM57468.1 KZS13953.1 GEFH01003519 JAP65062.1 GDIP01189571 JAJ33831.1 MNPL01015571 OQR71019.1 KU659968 AOG17766.1 ABLF02027290 GDIP01082329 JAM21386.1 GL732527 EFX87640.1 GDIP01199725 GDIQ01111462 JAJ23677.1 JAL40264.1 GDHC01019093 JAP99535.1 GDIP01187177 JAJ36225.1 GDIQ01148925 JAL02801.1 GBHO01021830 GBHO01015542 GBHO01001419 GDHC01005850 JAG21774.1 JAG28062.1 JAG42185.1 JAQ12779.1 GEFM01004115 JAP71681.1 GEGO01006784 JAR88620.1 KU659910 AOG17709.1 GBRD01002994 JAG62827.1 GDIP01138591 JAL65123.1 GDIP01175148 JAJ48254.1 GDIP01175147 JAJ48255.1 GDIP01140738 JAL62976.1 NCKU01005894 RWS04063.1 KQ424564 KOF70812.1 NEDP02076737 OWF35191.1 NCKU01005893 RWS04066.1 GFWV01006043 MAA30773.1 GFJQ02007381 JAV99588.1 RQTK01000003 RUS91969.1 KB203796 ESO83051.1 HACG01025300 CEK72165.1 GDIP01091824 JAM11891.1 AMQN01015980 KB312416 ELT87258.1 GDIQ01114170 JAL37556.1 GG666509 EEN60900.1 AADN05000878 GDIP01084031 JAM19684.1 JH816397 EKC40279.1 GDIP01085584 JAM18131.1 GDIP01009875 JAM93840.1
NWSH01000004 PCG81004.1 KQ459053 KPJ04167.1 GECU01004019 JAT03688.1 DS235756 EEB16444.1 GEZM01011792 JAV93436.1 NEVH01021928 PNF19301.1 KK852611 KDR20261.1 AJVK01014224 PYGN01000912 PSN39413.1 GEDC01012453 JAS24845.1 KQ971351 EFA06415.1 KK121013 KFM79549.1 UFQS01000041 UFQT01000041 SSW98306.1 SSX18692.1 MWRG01007578 PRD28170.1 KQ435851 KOX70831.1 CH480815 EDW43417.1 NNAY01002117 OXU22029.1 GL452770 EFN76801.1 KQ414617 KOC68203.1 KQ434899 KZC10952.1 LBMM01004397 KMQ92462.1 AAZX01012980 GFAC01004743 JAT94445.1 KK107274 QOIP01000008 EZA53732.1 RLU19300.1 GL763054 EFZ20284.1 GDKW01002553 JAI54042.1 KZ288291 PBC29210.1 JH431796 KQ976424 KYM88593.1 ADTU01024289 GECL01001245 JAP04879.1 GL437918 EFN69825.1 KQ977041 KYN06129.1 GBBI01002627 JAC16085.1 KQ978983 KYN26827.1 KQ982562 KYQ54925.1 GGMR01003574 MBY16193.1 CVRI01000059 CRL03231.1 GACK01005034 JAA60000.1 GL888275 EGI63445.1 GEDV01007903 JAP80654.1 KQ981993 KYN30903.1 GGMS01012712 MBY81915.1 GFPF01009949 MAA21095.1 GDIQ01010969 JAN83768.1 GBBM01003670 JAC31748.1 LJIG01016351 KRT80863.1 GDIP01046247 LRGB01001005 JAM57468.1 KZS13953.1 GEFH01003519 JAP65062.1 GDIP01189571 JAJ33831.1 MNPL01015571 OQR71019.1 KU659968 AOG17766.1 ABLF02027290 GDIP01082329 JAM21386.1 GL732527 EFX87640.1 GDIP01199725 GDIQ01111462 JAJ23677.1 JAL40264.1 GDHC01019093 JAP99535.1 GDIP01187177 JAJ36225.1 GDIQ01148925 JAL02801.1 GBHO01021830 GBHO01015542 GBHO01001419 GDHC01005850 JAG21774.1 JAG28062.1 JAG42185.1 JAQ12779.1 GEFM01004115 JAP71681.1 GEGO01006784 JAR88620.1 KU659910 AOG17709.1 GBRD01002994 JAG62827.1 GDIP01138591 JAL65123.1 GDIP01175148 JAJ48254.1 GDIP01175147 JAJ48255.1 GDIP01140738 JAL62976.1 NCKU01005894 RWS04063.1 KQ424564 KOF70812.1 NEDP02076737 OWF35191.1 NCKU01005893 RWS04066.1 GFWV01006043 MAA30773.1 GFJQ02007381 JAV99588.1 RQTK01000003 RUS91969.1 KB203796 ESO83051.1 HACG01025300 CEK72165.1 GDIP01091824 JAM11891.1 AMQN01015980 KB312416 ELT87258.1 GDIQ01114170 JAL37556.1 GG666509 EEN60900.1 AADN05000878 GDIP01084031 JAM19684.1 JH816397 EKC40279.1 GDIP01085584 JAM18131.1 GDIP01009875 JAM93840.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000009046
UP000235965
UP000027135
+ More
UP000092462 UP000192223 UP000245037 UP000007266 UP000054359 UP000053105 UP000001292 UP000215335 UP000008237 UP000053825 UP000076502 UP000036403 UP000002358 UP000053097 UP000279307 UP000005203 UP000242457 UP000078540 UP000005205 UP000000311 UP000078542 UP000078492 UP000075809 UP000183832 UP000007755 UP000078541 UP000076858 UP000192247 UP000007819 UP000000305 UP000285301 UP000053454 UP000242188 UP000271974 UP000030746 UP000085678 UP000014760 UP000001554 UP000000539 UP000005408 UP000189705
UP000092462 UP000192223 UP000245037 UP000007266 UP000054359 UP000053105 UP000001292 UP000215335 UP000008237 UP000053825 UP000076502 UP000036403 UP000002358 UP000053097 UP000279307 UP000005203 UP000242457 UP000078540 UP000005205 UP000000311 UP000078542 UP000078492 UP000075809 UP000183832 UP000007755 UP000078541 UP000076858 UP000192247 UP000007819 UP000000305 UP000285301 UP000053454 UP000242188 UP000271974 UP000030746 UP000085678 UP000014760 UP000001554 UP000000539 UP000005408 UP000189705
PRIDE
Interpro
SUPFAM
SSF53335
SSF53335
ProteinModelPortal
H9J4D4
A0A2A4KAR4
A0A194QFA7
A0A1B6JWU7
E0VST8
A0A1Y1NB44
+ More
A0A2J7PSI8 A0A067R9A5 A0A1B0DCU0 A0A1W4X4V1 A0A2P8Y580 A0A1B6DGV4 D6WRZ1 A0A087UQB0 A0A336K036 A0A2P6KPA9 A0A0N0BDR7 B4HGG4 A0A232EUI7 E2C5I8 A0A0L7RBI6 A0A154PGK9 A0A0J7KQ31 K7IWZ1 A0A1E1X5B7 A0A026WCD5 A0A087ZTT3 E9IGP1 A0A0P4VIY7 A0A2A3EBZ7 T1J262 A0A195BR29 A0A158NRS6 A0A0V0GBW6 E2A9M3 A0A195D150 A0A023F3T6 A0A195EG26 A0A151X3D4 A0A2S2NG63 A0A1J1ISQ6 L7M7H1 F4WQT2 A0A131YN03 A0A195ERQ2 A0A2S2QW06 A0A224YVY9 A0A0P6I1H8 A0A023GFK5 A0A0T6B077 A0A0N8DCV0 A0A131XFC5 A0A0P5B504 A0A1V9XBW1 A0A1B3PDH6 J9JK00 A0A0P5WTI7 E9FZ30 A0A0P5AFU4 A0A146KWJ1 A0A0N8A345 A0A0P5N6G7 A0A0A9Y4S1 A0A131Y1C5 A0A147BCX0 A0A1B3PDC1 A0A0K8TB88 A0A0N8CEV7 A0A0P5C603 A0A0P5C5P2 A0A0P5SFJ9 A0A3S3PLS7 A0A0L8G1B4 A0A210PFF4 A0A3S3NJR2 A0A293LPZ7 A0A1Z5KWW9 A0A3S1BYF3 V3ZQ95 A0A0B6ZWL5 A0A0P5W3U3 A0A1S3J0D7 R7T3J2 A0A0P5QIF2 C3YFD7 A0A1L1RJR6 A0A0N8CZC7 K1R9D4 A0A0P5WKY6 A0A1U7S5E4 A0A0P6BT66
A0A2J7PSI8 A0A067R9A5 A0A1B0DCU0 A0A1W4X4V1 A0A2P8Y580 A0A1B6DGV4 D6WRZ1 A0A087UQB0 A0A336K036 A0A2P6KPA9 A0A0N0BDR7 B4HGG4 A0A232EUI7 E2C5I8 A0A0L7RBI6 A0A154PGK9 A0A0J7KQ31 K7IWZ1 A0A1E1X5B7 A0A026WCD5 A0A087ZTT3 E9IGP1 A0A0P4VIY7 A0A2A3EBZ7 T1J262 A0A195BR29 A0A158NRS6 A0A0V0GBW6 E2A9M3 A0A195D150 A0A023F3T6 A0A195EG26 A0A151X3D4 A0A2S2NG63 A0A1J1ISQ6 L7M7H1 F4WQT2 A0A131YN03 A0A195ERQ2 A0A2S2QW06 A0A224YVY9 A0A0P6I1H8 A0A023GFK5 A0A0T6B077 A0A0N8DCV0 A0A131XFC5 A0A0P5B504 A0A1V9XBW1 A0A1B3PDH6 J9JK00 A0A0P5WTI7 E9FZ30 A0A0P5AFU4 A0A146KWJ1 A0A0N8A345 A0A0P5N6G7 A0A0A9Y4S1 A0A131Y1C5 A0A147BCX0 A0A1B3PDC1 A0A0K8TB88 A0A0N8CEV7 A0A0P5C603 A0A0P5C5P2 A0A0P5SFJ9 A0A3S3PLS7 A0A0L8G1B4 A0A210PFF4 A0A3S3NJR2 A0A293LPZ7 A0A1Z5KWW9 A0A3S1BYF3 V3ZQ95 A0A0B6ZWL5 A0A0P5W3U3 A0A1S3J0D7 R7T3J2 A0A0P5QIF2 C3YFD7 A0A1L1RJR6 A0A0N8CZC7 K1R9D4 A0A0P5WKY6 A0A1U7S5E4 A0A0P6BT66
PDB
5TEY
E-value=7.82626e-151,
Score=1370
Ontologies
GO
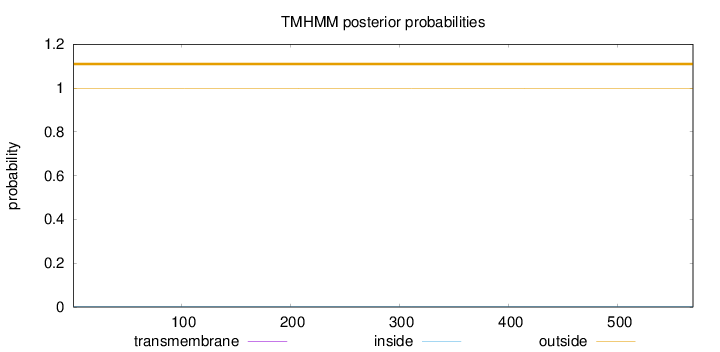
Topology
Length:
569
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000890000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00104
outside
1 - 569
Population Genetic Test Statistics
Pi
205.68121
Theta
160.95621
Tajima's D
1.36474
CLR
1.035877
CSRT
0.757162141892905
Interpretation
Uncertain
