Pre Gene Modal
BGIBMGA004489
Annotation
PREDICTED:_probable_enoyl-CoA_hydratase_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.718 Mitochondrial Reliability : 1.178
Sequence
CDS
ATGAGAATACTCATAAAGAAATATGCCACAATATCCAAAATTTTACTACCTAGACGAAATTTAGTCCGTTTCCTTTCAAATGATACGCAAAAAAAAACAGAAAATGAGACCGAAAAGATTGAAGAAGTAAAAAAAAATATCATAATAGATAAATTTGGAGCAGTCACAACATTGAACATAGACCGTCAAGTGACTCGCAACAGTCTAGATGAAGCCACCCTCAGGGAGATGGCTGCCGCAATAGATGACTTTGAGAAAGATAATCAAGCCAAAGTATTGGTTTTGAATGGTGAAGGAGGATCTTTCTGCTCTGGATTTGACATTGATGAAATTGGGGAGAAAGGCTATAACAGTTTAAAGGATGCTGCTGCACGTCTGTTACGTCGACCACTTTGTGATAAGCCAACAATAGCAGCCGTCACTGGATATGCAGTTGCGGAAGGTTTTGAGCTGGCATTAGCTTGTGATCTACGAGTCATTGAAGATACTGCAGTATTAGGATGTCTTGGAAGAAGATTTGGAGTCCCACTAAGCCTGTACGGTGCAAGGCGTTTGACATCATTGATTGGGTTGTCAAGAGCTCTCGATTTGATAATGACGGGGCGGCTAATCTCTGGAACTGATGCACAACAAATGGGCCTGGCTTGCAAACTTACTTCTACAGGAACCGGACTTGGTGAATCCGTGAAGTTAGCCAAATCCTTGGCTAAGTTTCCCCAAAATGCATTAATAATGGACAAGTTAGCAGCAGTTAACTCTCAGTTGAATCCGAACAGTGAGGAAAGCATGAGGGACGAAGCAGTCATGACTAGTCTGTTAGGTAACGCTATACTGGATATAGAGGAAGGCGTCAAAAAGTTTCAAGGAGGTATAGGAAAGCATGGAAAGTTCTACAAATTATCAGAAGCACCATTAAAAGAATGGGAAATTGAAGAGAGCGTCGAAGAAGTCACAGTTCAGAAGATAAATGAAAGGAAAAATGAAAAGTCATAA
Protein
MRILIKKYATISKILLPRRNLVRFLSNDTQKKTENETEKIEEVKKNIIIDKFGAVTTLNIDRQVTRNSLDEATLREMAAAIDDFEKDNQAKVLVLNGEGGSFCSGFDIDEIGEKGYNSLKDAAARLLRRPLCDKPTIAAVTGYAVAEGFELALACDLRVIEDTAVLGCLGRRFGVPLSLYGARRLTSLIGLSRALDLIMTGRLISGTDAQQMGLACKLTSTGTGLGESVKLAKSLAKFPQNALIMDKLAAVNSQLNPNSEESMRDEAVMTSLLGNAILDIEEGVKKFQGGIGKHGKFYKLSEAPLKEWEIEESVEEVTVQKINERKNEKS
Summary
Similarity
Belongs to the enoyl-CoA hydratase/isomerase family.
Uniprot
H9J4P9
A0A194QL57
A0A2A4JEQ0
A0A068FMP0
A0A2H1W7G3
A0A212F5Z0
+ More
D2XML3 A0A2W1BS93 A0A194QVX5 A0A293LM01 A0A2J7RA48 A0A067QV23 N6TUK2 U4UUQ2 A0A195C4W7 A0A1Z5KWE9 L7MIR5 A0A023FKQ3 V5IEB9 A0A131YND6 E9J8X0 A0A0C9RUB9 A0A1E1XFA5 E2BC82 A0A2L2YI43 B0X4F0 F4WBR4 J3JW70 A0A195FT01 E9GMS9 A0A151XHK6 A0A0J7L049 A0A195BD43 A0A3R7QY33 E2AXF1 A0A0P5N3S8 A0A0P6B028 A0A0N8BVV9 A0A0P6I6G1 A0A0P5M3F7 A0A0P5FSE2 A0A158NRG3 U5EW82 A0A154PDA4 A0A0N8ARK6 A0A0P5BVK2 A0A1Y1JRB2 A0A182LWG9 A0A1Y3B3A5 A0A132A7R0 A0A226ENR9 A0A182G921 A0A1Q3F3C6 L7MG18 A0A2T7PGF5 A0A182HCB0 A0A1B0CEY3 A0A1W4W7E1 A0A182VZL8 A0A182TW67 A0A0K8TM54 A0A1D2NI52 A0A182V0Q9 A0A0N7ZB84 A0A182WST0 A0A0C9QHP4 A0A182RZM4 A0A0L7RGB9 A0A1W0WTX9 A0A182HLN3 D6W8S7 A0A1S4F3Y4 A0A0P4W4R6 A0A182QHY8 Q17G32 A0A3F2YUU2 Q7PY24 Q4JSB0 A0A182JMU8 A0A182Y3S7 A0A182NRU0 A0A026WKU8 A0A023ENS5 A0A182PSJ1 A0A182K5W7 A0A182SXL2 A0A1D1VDV8 V3ZYN5 A0A1A9WPG8 A0A2A3E2W3 A0A0L0CIM3 A0A0L8GZN0 A0A1Q3LIN8 A0A1B6MHD7 W5J4F3 A0A0V1FMX8
D2XML3 A0A2W1BS93 A0A194QVX5 A0A293LM01 A0A2J7RA48 A0A067QV23 N6TUK2 U4UUQ2 A0A195C4W7 A0A1Z5KWE9 L7MIR5 A0A023FKQ3 V5IEB9 A0A131YND6 E9J8X0 A0A0C9RUB9 A0A1E1XFA5 E2BC82 A0A2L2YI43 B0X4F0 F4WBR4 J3JW70 A0A195FT01 E9GMS9 A0A151XHK6 A0A0J7L049 A0A195BD43 A0A3R7QY33 E2AXF1 A0A0P5N3S8 A0A0P6B028 A0A0N8BVV9 A0A0P6I6G1 A0A0P5M3F7 A0A0P5FSE2 A0A158NRG3 U5EW82 A0A154PDA4 A0A0N8ARK6 A0A0P5BVK2 A0A1Y1JRB2 A0A182LWG9 A0A1Y3B3A5 A0A132A7R0 A0A226ENR9 A0A182G921 A0A1Q3F3C6 L7MG18 A0A2T7PGF5 A0A182HCB0 A0A1B0CEY3 A0A1W4W7E1 A0A182VZL8 A0A182TW67 A0A0K8TM54 A0A1D2NI52 A0A182V0Q9 A0A0N7ZB84 A0A182WST0 A0A0C9QHP4 A0A182RZM4 A0A0L7RGB9 A0A1W0WTX9 A0A182HLN3 D6W8S7 A0A1S4F3Y4 A0A0P4W4R6 A0A182QHY8 Q17G32 A0A3F2YUU2 Q7PY24 Q4JSB0 A0A182JMU8 A0A182Y3S7 A0A182NRU0 A0A026WKU8 A0A023ENS5 A0A182PSJ1 A0A182K5W7 A0A182SXL2 A0A1D1VDV8 V3ZYN5 A0A1A9WPG8 A0A2A3E2W3 A0A0L0CIM3 A0A0L8GZN0 A0A1Q3LIN8 A0A1B6MHD7 W5J4F3 A0A0V1FMX8
Pubmed
19121390
26354079
26385554
22118469
20074338
28756777
+ More
24845553 23537049 28528879 25576852 25765539 26830274 21282665 26131772 28503490 20798317 26561354 21719571 22516182 21292972 21347285 28004739 26555130 26483478 26369729 27289101 18362917 19820115 17510324 20966253 12364791 14747013 17210077 15944077 25244985 24508170 30249741 24945155 27649274 23254933 26108605 20920257 23761445
24845553 23537049 28528879 25576852 25765539 26830274 21282665 26131772 28503490 20798317 26561354 21719571 22516182 21292972 21347285 28004739 26555130 26483478 26369729 27289101 18362917 19820115 17510324 20966253 12364791 14747013 17210077 15944077 25244985 24508170 30249741 24945155 27649274 23254933 26108605 20920257 23761445
EMBL
BABH01022070
KQ459053
KPJ04176.1
NWSH01001732
PCG70299.1
KJ622096
+ More
AID66686.1 ODYU01006507 SOQ48414.1 AGBW02010110 OWR49155.1 GU205159 ADB57046.1 KZ149954 PZC76514.1 KQ461181 KPJ07716.1 GFWV01003130 MAA27860.1 NEVH01006578 PNF37700.1 KK853353 KDR08205.1 APGK01051698 APGK01051699 KB741201 ENN72955.1 KB632375 ERL93880.1 KQ978317 KYM95203.1 GFJQ02007776 JAV99193.1 GACK01001866 JAA63168.1 GBBK01003234 JAC21248.1 GANP01010457 JAB74011.1 GEDV01007788 JAP80769.1 GL769162 EFZ10718.1 GBZX01001744 JAG90996.1 GFAC01001310 JAT97878.1 GL447256 EFN86709.1 IAAA01029450 LAA07779.1 DS232338 EDS40303.1 GL888066 EGI68342.1 BT127488 AEE62450.1 KQ981280 KYN43427.1 GL732553 EFX79236.1 KQ982123 KYQ59886.1 LBMM01001522 KMQ96152.1 KQ976511 KYM82491.1 QCYY01000779 ROT82744.1 GL443548 EFN61931.1 GDIQ01147441 JAL04285.1 GDIP01021936 LRGB01002384 JAM81779.1 KZS07865.1 GDIQ01139787 JAL11939.1 GDIQ01008737 JAN86000.1 GDIQ01161537 JAK90188.1 GDIQ01254012 JAJ97712.1 ADTU01023964 GANO01001546 JAB58325.1 KQ434878 KZC09885.1 GDIQ01249811 JAK01914.1 GDIP01179307 JAJ44095.1 GEZM01102889 JAV51744.1 AXCM01000026 MUJZ01047581 OTF74343.1 JXLN01011022 KPM06675.1 LNIX01000003 OXA58664.1 JXUM01008327 KQ560258 KXJ83508.1 GFDL01012985 JAV22060.1 GACK01001893 JAA63141.1 PZQS01000004 PVD32497.1 JXUM01033300 KQ560983 KXJ80131.1 AJWK01009463 GDAI01002392 JAI15211.1 LJIJ01000036 ODN04765.1 GDRN01088036 JAI60936.1 GBYB01003054 JAG72821.1 KQ414596 KOC69900.1 MTYJ01000047 OQV18651.1 APCN01000909 KQ971312 EEZ98266.1 GDRN01088040 GDRN01088038 JAI60934.1 AXCN02001148 CH477266 EAT45553.1 AAAB01008987 EAA00852.3 CR954257 CAJ14152.1 KK107159 QOIP01000001 EZA56672.1 RLU27083.1 GAPW01002511 JAC11087.1 BDGG01000005 GAU99080.1 KB203188 ESO86101.1 KZ288416 PBC26040.1 JRES01000328 KNC32258.1 KQ419775 KOF82402.1 MKRV01000045 OJU45191.1 GEBQ01004620 JAT35357.1 ADMH02002143 ETN58293.1 JYDT01000057 KRY87324.1
AID66686.1 ODYU01006507 SOQ48414.1 AGBW02010110 OWR49155.1 GU205159 ADB57046.1 KZ149954 PZC76514.1 KQ461181 KPJ07716.1 GFWV01003130 MAA27860.1 NEVH01006578 PNF37700.1 KK853353 KDR08205.1 APGK01051698 APGK01051699 KB741201 ENN72955.1 KB632375 ERL93880.1 KQ978317 KYM95203.1 GFJQ02007776 JAV99193.1 GACK01001866 JAA63168.1 GBBK01003234 JAC21248.1 GANP01010457 JAB74011.1 GEDV01007788 JAP80769.1 GL769162 EFZ10718.1 GBZX01001744 JAG90996.1 GFAC01001310 JAT97878.1 GL447256 EFN86709.1 IAAA01029450 LAA07779.1 DS232338 EDS40303.1 GL888066 EGI68342.1 BT127488 AEE62450.1 KQ981280 KYN43427.1 GL732553 EFX79236.1 KQ982123 KYQ59886.1 LBMM01001522 KMQ96152.1 KQ976511 KYM82491.1 QCYY01000779 ROT82744.1 GL443548 EFN61931.1 GDIQ01147441 JAL04285.1 GDIP01021936 LRGB01002384 JAM81779.1 KZS07865.1 GDIQ01139787 JAL11939.1 GDIQ01008737 JAN86000.1 GDIQ01161537 JAK90188.1 GDIQ01254012 JAJ97712.1 ADTU01023964 GANO01001546 JAB58325.1 KQ434878 KZC09885.1 GDIQ01249811 JAK01914.1 GDIP01179307 JAJ44095.1 GEZM01102889 JAV51744.1 AXCM01000026 MUJZ01047581 OTF74343.1 JXLN01011022 KPM06675.1 LNIX01000003 OXA58664.1 JXUM01008327 KQ560258 KXJ83508.1 GFDL01012985 JAV22060.1 GACK01001893 JAA63141.1 PZQS01000004 PVD32497.1 JXUM01033300 KQ560983 KXJ80131.1 AJWK01009463 GDAI01002392 JAI15211.1 LJIJ01000036 ODN04765.1 GDRN01088036 JAI60936.1 GBYB01003054 JAG72821.1 KQ414596 KOC69900.1 MTYJ01000047 OQV18651.1 APCN01000909 KQ971312 EEZ98266.1 GDRN01088040 GDRN01088038 JAI60934.1 AXCN02001148 CH477266 EAT45553.1 AAAB01008987 EAA00852.3 CR954257 CAJ14152.1 KK107159 QOIP01000001 EZA56672.1 RLU27083.1 GAPW01002511 JAC11087.1 BDGG01000005 GAU99080.1 KB203188 ESO86101.1 KZ288416 PBC26040.1 JRES01000328 KNC32258.1 KQ419775 KOF82402.1 MKRV01000045 OJU45191.1 GEBQ01004620 JAT35357.1 ADMH02002143 ETN58293.1 JYDT01000057 KRY87324.1
Proteomes
UP000005204
UP000053268
UP000218220
UP000007151
UP000053240
UP000235965
+ More
UP000027135 UP000019118 UP000030742 UP000078542 UP000008237 UP000002320 UP000007755 UP000078541 UP000000305 UP000075809 UP000036403 UP000078540 UP000283509 UP000000311 UP000076858 UP000005205 UP000076502 UP000075883 UP000198287 UP000069940 UP000249989 UP000245119 UP000092461 UP000192223 UP000075920 UP000075902 UP000094527 UP000075903 UP000076407 UP000075900 UP000053825 UP000075840 UP000007266 UP000075886 UP000008820 UP000075882 UP000007062 UP000075880 UP000076408 UP000075884 UP000053097 UP000279307 UP000075885 UP000075881 UP000075901 UP000186922 UP000030746 UP000091820 UP000242457 UP000037069 UP000053454 UP000000673 UP000054995
UP000027135 UP000019118 UP000030742 UP000078542 UP000008237 UP000002320 UP000007755 UP000078541 UP000000305 UP000075809 UP000036403 UP000078540 UP000283509 UP000000311 UP000076858 UP000005205 UP000076502 UP000075883 UP000198287 UP000069940 UP000249989 UP000245119 UP000092461 UP000192223 UP000075920 UP000075902 UP000094527 UP000075903 UP000076407 UP000075900 UP000053825 UP000075840 UP000007266 UP000075886 UP000008820 UP000075882 UP000007062 UP000075880 UP000076408 UP000075884 UP000053097 UP000279307 UP000075885 UP000075881 UP000075901 UP000186922 UP000030746 UP000091820 UP000242457 UP000037069 UP000053454 UP000000673 UP000054995
PRIDE
Pfam
PF00378 ECH_1
Interpro
SUPFAM
SSF52096
SSF52096
ProteinModelPortal
H9J4P9
A0A194QL57
A0A2A4JEQ0
A0A068FMP0
A0A2H1W7G3
A0A212F5Z0
+ More
D2XML3 A0A2W1BS93 A0A194QVX5 A0A293LM01 A0A2J7RA48 A0A067QV23 N6TUK2 U4UUQ2 A0A195C4W7 A0A1Z5KWE9 L7MIR5 A0A023FKQ3 V5IEB9 A0A131YND6 E9J8X0 A0A0C9RUB9 A0A1E1XFA5 E2BC82 A0A2L2YI43 B0X4F0 F4WBR4 J3JW70 A0A195FT01 E9GMS9 A0A151XHK6 A0A0J7L049 A0A195BD43 A0A3R7QY33 E2AXF1 A0A0P5N3S8 A0A0P6B028 A0A0N8BVV9 A0A0P6I6G1 A0A0P5M3F7 A0A0P5FSE2 A0A158NRG3 U5EW82 A0A154PDA4 A0A0N8ARK6 A0A0P5BVK2 A0A1Y1JRB2 A0A182LWG9 A0A1Y3B3A5 A0A132A7R0 A0A226ENR9 A0A182G921 A0A1Q3F3C6 L7MG18 A0A2T7PGF5 A0A182HCB0 A0A1B0CEY3 A0A1W4W7E1 A0A182VZL8 A0A182TW67 A0A0K8TM54 A0A1D2NI52 A0A182V0Q9 A0A0N7ZB84 A0A182WST0 A0A0C9QHP4 A0A182RZM4 A0A0L7RGB9 A0A1W0WTX9 A0A182HLN3 D6W8S7 A0A1S4F3Y4 A0A0P4W4R6 A0A182QHY8 Q17G32 A0A3F2YUU2 Q7PY24 Q4JSB0 A0A182JMU8 A0A182Y3S7 A0A182NRU0 A0A026WKU8 A0A023ENS5 A0A182PSJ1 A0A182K5W7 A0A182SXL2 A0A1D1VDV8 V3ZYN5 A0A1A9WPG8 A0A2A3E2W3 A0A0L0CIM3 A0A0L8GZN0 A0A1Q3LIN8 A0A1B6MHD7 W5J4F3 A0A0V1FMX8
D2XML3 A0A2W1BS93 A0A194QVX5 A0A293LM01 A0A2J7RA48 A0A067QV23 N6TUK2 U4UUQ2 A0A195C4W7 A0A1Z5KWE9 L7MIR5 A0A023FKQ3 V5IEB9 A0A131YND6 E9J8X0 A0A0C9RUB9 A0A1E1XFA5 E2BC82 A0A2L2YI43 B0X4F0 F4WBR4 J3JW70 A0A195FT01 E9GMS9 A0A151XHK6 A0A0J7L049 A0A195BD43 A0A3R7QY33 E2AXF1 A0A0P5N3S8 A0A0P6B028 A0A0N8BVV9 A0A0P6I6G1 A0A0P5M3F7 A0A0P5FSE2 A0A158NRG3 U5EW82 A0A154PDA4 A0A0N8ARK6 A0A0P5BVK2 A0A1Y1JRB2 A0A182LWG9 A0A1Y3B3A5 A0A132A7R0 A0A226ENR9 A0A182G921 A0A1Q3F3C6 L7MG18 A0A2T7PGF5 A0A182HCB0 A0A1B0CEY3 A0A1W4W7E1 A0A182VZL8 A0A182TW67 A0A0K8TM54 A0A1D2NI52 A0A182V0Q9 A0A0N7ZB84 A0A182WST0 A0A0C9QHP4 A0A182RZM4 A0A0L7RGB9 A0A1W0WTX9 A0A182HLN3 D6W8S7 A0A1S4F3Y4 A0A0P4W4R6 A0A182QHY8 Q17G32 A0A3F2YUU2 Q7PY24 Q4JSB0 A0A182JMU8 A0A182Y3S7 A0A182NRU0 A0A026WKU8 A0A023ENS5 A0A182PSJ1 A0A182K5W7 A0A182SXL2 A0A1D1VDV8 V3ZYN5 A0A1A9WPG8 A0A2A3E2W3 A0A0L0CIM3 A0A0L8GZN0 A0A1Q3LIN8 A0A1B6MHD7 W5J4F3 A0A0V1FMX8
PDB
4Z0M
E-value=1.01055e-38,
Score=401
Ontologies
GO
Topology
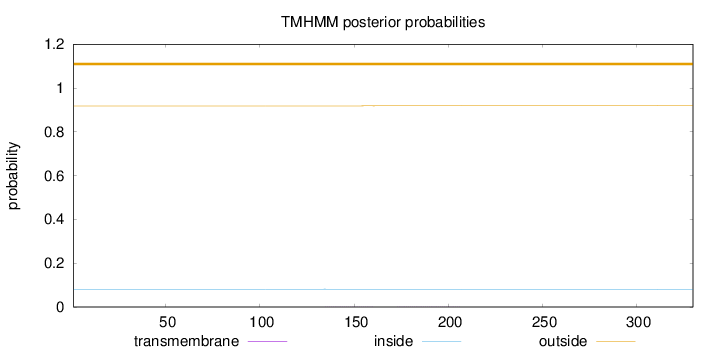
Length:
330
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02549
Exp number, first 60 AAs:
0
Total prob of N-in:
0.08150
outside
1 - 330
Population Genetic Test Statistics
Pi
206.730162
Theta
179.746407
Tajima's D
0.45202
CLR
32.029932
CSRT
0.493175341232938
Interpretation
Uncertain
