Pre Gene Modal
BGIBMGA004377
Annotation
Der1-like_domain_family_member_1_[Bombyx_mori]
Full name
Derlin
Location in the cell
PlasmaMembrane Reliability : 4.638
Sequence
CDS
ATGTCGGAATTCCGAGATTGGTACAATGGTGTACCGTTTTTCACCAGATATTGGATGACTTTTACTATTGTATTGAGTTTGTTTGGTAAATTTGGGCTTGTGAGCCCTTATTATTTTATACTGGATTTCTATTATTTCTTCAATCAATTTCAGATATGGCGTCCATTAACAGCTTTGTTTTACTATCCAATAAATCCAGGCACAGGATTTCATTTCCTGATAAACTGTTACTTTTTGTACAACTACTCTCAAAGACTTGAGACCGGCATGTTTGCAGGGAAGCCAGCTGACTATTTCTACATGCTTTTGTTTAATTGGGTGTGCTGTGTTATCATTGGATTACTTGTCAAATTGCCAGTATTAATGGACCCGATGGTACTCTCGGTATTATATGTATGGTGTCAGCTTAACAAAGATGTAATTGTATCATTTTGGTTTGGCACAAGATTTAAGGCTATGTATTTACCTTGGGTACTACTCGCTTTCAACCTTGTCATCAGTGGAGGTGGGATAATGGAGCTCCTAGGCATTCTAATTGGTCACTTAGCATTCTTCCTTTTGTTCAAATACCCCCAAGAGTTTGGTGGCCCTGCATTACTCACACCACCAGCATTCTTGAAACAACTATTCCCGGACACTAGGTACGTGGGTGGTTTTGGTACAGCGCCACAAGCGAGAGTACCCACTCGCCCAGGCAACACCGTGTTCGGAGGACATAACTGGGGCAGAGGACAAACTCTTGGAGGAAACTGA
Protein
MSEFRDWYNGVPFFTRYWMTFTIVLSLFGKFGLVSPYYFILDFYYFFNQFQIWRPLTALFYYPINPGTGFHFLINCYFLYNYSQRLETGMFAGKPADYFYMLLFNWVCCVIIGLLVKLPVLMDPMVLSVLYVWCQLNKDVIVSFWFGTRFKAMYLPWVLLAFNLVISGGGIMELLGILIGHLAFFLLFKYPQEFGGPALLTPPAFLKQLFPDTRYVGGFGTAPQARVPTRPGNTVFGGHNWGRGQTLGGN
Summary
Description
May be involved in the degradation of misfolded endoplasmic reticulum (ER) luminal proteins.
Similarity
Belongs to the derlin family.
Uniprot
H9J4D8
Q2F6B5
A0A1E1W8Z3
A0A2A4JEB5
A0A2W1BXL7
I4DQY9
+ More
A0A194Q5G7 A0A212F5Z7 S4PJW6 A0A0N1IGV5 U5EZG9 E9FQR1 A0A0P5U053 A0A0P6ERC4 D6WPF8 A0A0J7K3J6 A0A154NZS9 A0A1Y1KU85 A0A1W4X2Z2 A0A151IIF4 A0A026WJX5 F4WNI4 C4WWH3 A0A195FXL8 E2A1L2 A0A0M8ZRB1 A0A151WVJ3 A0A0L7QP74 J9JN94 A0A2P8YSR7 A0A2H8TIU3 A0A2M3ZHU3 A0A0P6BNB9 A0A2S2NM81 V9IJT8 A0A088A6M4 A0A2M4AG83 A0A0P6GVJ4 T1DLD5 A0A2M4BX90 A0A1B6E1M1 A0A195B891 A0A195ED21 A0A0P5AS76 A0A0P5MDF2 Q1HQM9 Q172Y9 A0A1S3DSK7 A0A1D1V639 W5JWC6 A0A1B6LVL5 A0A1B6GVF9 A0A0R3RZ64 A0A182E5H0 A0A2K6VFR7 A0A0B2UU68 W4YQI8 A0A0V0G5Z3 K1R606 A0A0P4VL95 A0A0N5CX61 A0A0P5IGJ1 A0A182TGY3 A0A182MR07 A0A1B6HKV4 A0A0B7BDI0 A0A0N8D1T5 V4B2N8 A0A023EJV1 A0A1S4H1G9 A0A2C9K2T2 A0A3P6UIP3 A0A2S2R0H3 A0A1C7CZP8 A0A182K639 A0A067RCR3 A0A3S1BC59 A0A1I7VXX0 A0A0A9W3J1 A0A182YP94 N6TN55 Q7PVG0 A0A183XCV6 A0A182VL38 A0A0L8GR20 U4UH75 A0A182IJ79 F7ASI0 A0A1S3KBN5 A0A210QQN9 A0A182X0R3 A0A182HGF0 C3YFB1 H2Z1B1 J3JV97 A0A0I9N9F4 A0A182SLZ2 A0A182PG21 A0A0N4TWP8
A0A194Q5G7 A0A212F5Z7 S4PJW6 A0A0N1IGV5 U5EZG9 E9FQR1 A0A0P5U053 A0A0P6ERC4 D6WPF8 A0A0J7K3J6 A0A154NZS9 A0A1Y1KU85 A0A1W4X2Z2 A0A151IIF4 A0A026WJX5 F4WNI4 C4WWH3 A0A195FXL8 E2A1L2 A0A0M8ZRB1 A0A151WVJ3 A0A0L7QP74 J9JN94 A0A2P8YSR7 A0A2H8TIU3 A0A2M3ZHU3 A0A0P6BNB9 A0A2S2NM81 V9IJT8 A0A088A6M4 A0A2M4AG83 A0A0P6GVJ4 T1DLD5 A0A2M4BX90 A0A1B6E1M1 A0A195B891 A0A195ED21 A0A0P5AS76 A0A0P5MDF2 Q1HQM9 Q172Y9 A0A1S3DSK7 A0A1D1V639 W5JWC6 A0A1B6LVL5 A0A1B6GVF9 A0A0R3RZ64 A0A182E5H0 A0A2K6VFR7 A0A0B2UU68 W4YQI8 A0A0V0G5Z3 K1R606 A0A0P4VL95 A0A0N5CX61 A0A0P5IGJ1 A0A182TGY3 A0A182MR07 A0A1B6HKV4 A0A0B7BDI0 A0A0N8D1T5 V4B2N8 A0A023EJV1 A0A1S4H1G9 A0A2C9K2T2 A0A3P6UIP3 A0A2S2R0H3 A0A1C7CZP8 A0A182K639 A0A067RCR3 A0A3S1BC59 A0A1I7VXX0 A0A0A9W3J1 A0A182YP94 N6TN55 Q7PVG0 A0A183XCV6 A0A182VL38 A0A0L8GR20 U4UH75 A0A182IJ79 F7ASI0 A0A1S3KBN5 A0A210QQN9 A0A182X0R3 A0A182HGF0 C3YFB1 H2Z1B1 J3JV97 A0A0I9N9F4 A0A182SLZ2 A0A182PG21 A0A0N4TWP8
Pubmed
19121390
28756777
22651552
26354079
22118469
23622113
+ More
21292972 18362917 19820115 28004739 24508170 30249741 21719571 20798317 29403074 17204158 17510324 27649274 20920257 23761445 22919073 22992520 27129103 23254933 24945155 12364791 15562597 20966253 24845553 25217238 25401762 26823975 25244985 23537049 26850696 12481130 15114417 28812685 18563158 22516182 17885136
21292972 18362917 19820115 28004739 24508170 30249741 21719571 20798317 29403074 17204158 17510324 27649274 20920257 23761445 22919073 22992520 27129103 23254933 24945155 12364791 15562597 20966253 24845553 25217238 25401762 26823975 25244985 23537049 26850696 12481130 15114417 28812685 18563158 22516182 17885136
EMBL
BABH01022069
DQ311157
ABD36102.1
GDQN01007611
GDQN01006678
JAT83443.1
+ More
JAT84376.1 NWSH01001732 PCG70301.1 KZ149954 PZC76513.1 AK404849 BAM20329.1 KQ459460 KPJ00788.1 AGBW02010110 OWR49156.1 GAIX01004785 JAA87775.1 KQ459838 KPJ19732.1 GANO01000819 JAB59052.1 GL732523 EFX90326.1 GDIP01119658 LRGB01002568 JAL84056.1 KZS07108.1 GDIQ01072378 JAN22359.1 KQ971354 EFA06805.1 LBMM01015210 KMQ84889.1 KQ434786 KZC05175.1 GEZM01077523 JAV63285.1 KQ977477 KYN02456.1 KK107167 QOIP01000010 EZA56352.1 RLU18106.1 GL888237 EGI64177.1 AK342065 BAH72243.1 KQ981215 KYN44589.1 GL435766 EFN72685.1 KQ435955 KOX68043.1 KQ982706 KYQ51858.1 KQ414843 KOC60301.1 ABLF02005025 PYGN01000381 PSN47291.1 GFXV01002164 MBW13969.1 GGFM01007328 MBW28079.1 GDIP01012547 JAM91168.1 GGMR01005618 MBY18237.1 JR048749 AEY60751.1 GGFK01006479 MBW39800.1 GDIQ01027819 JAN66918.1 GAMD01000506 JAB01085.1 GGFJ01008555 MBW57696.1 GEDC01030786 GEDC01005494 JAS06512.1 JAS31804.1 KQ976565 KYM80465.1 KQ979074 KYN22717.1 GDIP01209203 JAJ14199.1 GDIQ01161340 JAK90385.1 DQ440415 ABF18448.1 CH477429 EAT41090.1 BDGG01000003 GAU96340.1 ADMH02000176 ETN67500.1 GEBQ01012246 JAT27731.1 GECZ01003436 JAS66333.1 UYRW01000601 VDK68447.1 CMVM020000024 JPKZ01003226 KHN72637.1 AAGJ04146733 GECL01003012 JAP03112.1 JH823229 EKC29406.1 GDKW01001420 JAI55175.1 UYYF01004313 VDN02149.1 GDIQ01217319 JAK34406.1 AXCM01004997 GECU01032394 JAS75312.1 HACG01044153 CEK91018.1 GDIP01077167 JAM26548.1 KB203854 ESO82769.1 GAPW01004268 JAC09330.1 AAAB01008984 UYRX01000270 VDK79008.1 GGMS01013679 MBY82882.1 KK852722 KDR17716.1 RQTK01000411 RUS80103.1 JH712250 EFO25036.1 GBHO01041623 GBRD01013198 GDHC01000253 JAG01981.1 JAG52628.1 JAQ18376.1 APGK01058320 KB741288 ENN70655.1 EAA14829.4 UYWW01012185 VDM19424.1 KQ420813 KOF79269.1 KB632322 ERL92357.1 NEDP02002396 OWF51050.1 APCN01004219 GG666509 EEN60874.1 BT127164 AEE62126.1 LN857024 CTP81790.1 UZAD01013373 VDN94479.1
JAT84376.1 NWSH01001732 PCG70301.1 KZ149954 PZC76513.1 AK404849 BAM20329.1 KQ459460 KPJ00788.1 AGBW02010110 OWR49156.1 GAIX01004785 JAA87775.1 KQ459838 KPJ19732.1 GANO01000819 JAB59052.1 GL732523 EFX90326.1 GDIP01119658 LRGB01002568 JAL84056.1 KZS07108.1 GDIQ01072378 JAN22359.1 KQ971354 EFA06805.1 LBMM01015210 KMQ84889.1 KQ434786 KZC05175.1 GEZM01077523 JAV63285.1 KQ977477 KYN02456.1 KK107167 QOIP01000010 EZA56352.1 RLU18106.1 GL888237 EGI64177.1 AK342065 BAH72243.1 KQ981215 KYN44589.1 GL435766 EFN72685.1 KQ435955 KOX68043.1 KQ982706 KYQ51858.1 KQ414843 KOC60301.1 ABLF02005025 PYGN01000381 PSN47291.1 GFXV01002164 MBW13969.1 GGFM01007328 MBW28079.1 GDIP01012547 JAM91168.1 GGMR01005618 MBY18237.1 JR048749 AEY60751.1 GGFK01006479 MBW39800.1 GDIQ01027819 JAN66918.1 GAMD01000506 JAB01085.1 GGFJ01008555 MBW57696.1 GEDC01030786 GEDC01005494 JAS06512.1 JAS31804.1 KQ976565 KYM80465.1 KQ979074 KYN22717.1 GDIP01209203 JAJ14199.1 GDIQ01161340 JAK90385.1 DQ440415 ABF18448.1 CH477429 EAT41090.1 BDGG01000003 GAU96340.1 ADMH02000176 ETN67500.1 GEBQ01012246 JAT27731.1 GECZ01003436 JAS66333.1 UYRW01000601 VDK68447.1 CMVM020000024 JPKZ01003226 KHN72637.1 AAGJ04146733 GECL01003012 JAP03112.1 JH823229 EKC29406.1 GDKW01001420 JAI55175.1 UYYF01004313 VDN02149.1 GDIQ01217319 JAK34406.1 AXCM01004997 GECU01032394 JAS75312.1 HACG01044153 CEK91018.1 GDIP01077167 JAM26548.1 KB203854 ESO82769.1 GAPW01004268 JAC09330.1 AAAB01008984 UYRX01000270 VDK79008.1 GGMS01013679 MBY82882.1 KK852722 KDR17716.1 RQTK01000411 RUS80103.1 JH712250 EFO25036.1 GBHO01041623 GBRD01013198 GDHC01000253 JAG01981.1 JAG52628.1 JAQ18376.1 APGK01058320 KB741288 ENN70655.1 EAA14829.4 UYWW01012185 VDM19424.1 KQ420813 KOF79269.1 KB632322 ERL92357.1 NEDP02002396 OWF51050.1 APCN01004219 GG666509 EEN60874.1 BT127164 AEE62126.1 LN857024 CTP81790.1 UZAD01013373 VDN94479.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000000305
+ More
UP000076858 UP000007266 UP000036403 UP000076502 UP000192223 UP000078542 UP000053097 UP000279307 UP000007755 UP000078541 UP000000311 UP000053105 UP000075809 UP000053825 UP000007819 UP000245037 UP000005203 UP000078540 UP000078492 UP000008820 UP000079169 UP000186922 UP000000673 UP000050640 UP000077448 UP000271087 UP000024404 UP000031036 UP000007110 UP000005408 UP000046394 UP000276776 UP000075902 UP000075883 UP000030746 UP000076420 UP000277928 UP000075882 UP000075881 UP000027135 UP000271974 UP000095285 UP000076408 UP000019118 UP000007062 UP000093561 UP000270924 UP000075903 UP000053454 UP000030742 UP000075880 UP000008144 UP000085678 UP000242188 UP000076407 UP000075840 UP000001554 UP000007875 UP000006672 UP000075901 UP000075885 UP000038020 UP000278627
UP000076858 UP000007266 UP000036403 UP000076502 UP000192223 UP000078542 UP000053097 UP000279307 UP000007755 UP000078541 UP000000311 UP000053105 UP000075809 UP000053825 UP000007819 UP000245037 UP000005203 UP000078540 UP000078492 UP000008820 UP000079169 UP000186922 UP000000673 UP000050640 UP000077448 UP000271087 UP000024404 UP000031036 UP000007110 UP000005408 UP000046394 UP000276776 UP000075902 UP000075883 UP000030746 UP000076420 UP000277928 UP000075882 UP000075881 UP000027135 UP000271974 UP000095285 UP000076408 UP000019118 UP000007062 UP000093561 UP000270924 UP000075903 UP000053454 UP000030742 UP000075880 UP000008144 UP000085678 UP000242188 UP000076407 UP000075840 UP000001554 UP000007875 UP000006672 UP000075901 UP000075885 UP000038020 UP000278627
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9J4D8
Q2F6B5
A0A1E1W8Z3
A0A2A4JEB5
A0A2W1BXL7
I4DQY9
+ More
A0A194Q5G7 A0A212F5Z7 S4PJW6 A0A0N1IGV5 U5EZG9 E9FQR1 A0A0P5U053 A0A0P6ERC4 D6WPF8 A0A0J7K3J6 A0A154NZS9 A0A1Y1KU85 A0A1W4X2Z2 A0A151IIF4 A0A026WJX5 F4WNI4 C4WWH3 A0A195FXL8 E2A1L2 A0A0M8ZRB1 A0A151WVJ3 A0A0L7QP74 J9JN94 A0A2P8YSR7 A0A2H8TIU3 A0A2M3ZHU3 A0A0P6BNB9 A0A2S2NM81 V9IJT8 A0A088A6M4 A0A2M4AG83 A0A0P6GVJ4 T1DLD5 A0A2M4BX90 A0A1B6E1M1 A0A195B891 A0A195ED21 A0A0P5AS76 A0A0P5MDF2 Q1HQM9 Q172Y9 A0A1S3DSK7 A0A1D1V639 W5JWC6 A0A1B6LVL5 A0A1B6GVF9 A0A0R3RZ64 A0A182E5H0 A0A2K6VFR7 A0A0B2UU68 W4YQI8 A0A0V0G5Z3 K1R606 A0A0P4VL95 A0A0N5CX61 A0A0P5IGJ1 A0A182TGY3 A0A182MR07 A0A1B6HKV4 A0A0B7BDI0 A0A0N8D1T5 V4B2N8 A0A023EJV1 A0A1S4H1G9 A0A2C9K2T2 A0A3P6UIP3 A0A2S2R0H3 A0A1C7CZP8 A0A182K639 A0A067RCR3 A0A3S1BC59 A0A1I7VXX0 A0A0A9W3J1 A0A182YP94 N6TN55 Q7PVG0 A0A183XCV6 A0A182VL38 A0A0L8GR20 U4UH75 A0A182IJ79 F7ASI0 A0A1S3KBN5 A0A210QQN9 A0A182X0R3 A0A182HGF0 C3YFB1 H2Z1B1 J3JV97 A0A0I9N9F4 A0A182SLZ2 A0A182PG21 A0A0N4TWP8
A0A194Q5G7 A0A212F5Z7 S4PJW6 A0A0N1IGV5 U5EZG9 E9FQR1 A0A0P5U053 A0A0P6ERC4 D6WPF8 A0A0J7K3J6 A0A154NZS9 A0A1Y1KU85 A0A1W4X2Z2 A0A151IIF4 A0A026WJX5 F4WNI4 C4WWH3 A0A195FXL8 E2A1L2 A0A0M8ZRB1 A0A151WVJ3 A0A0L7QP74 J9JN94 A0A2P8YSR7 A0A2H8TIU3 A0A2M3ZHU3 A0A0P6BNB9 A0A2S2NM81 V9IJT8 A0A088A6M4 A0A2M4AG83 A0A0P6GVJ4 T1DLD5 A0A2M4BX90 A0A1B6E1M1 A0A195B891 A0A195ED21 A0A0P5AS76 A0A0P5MDF2 Q1HQM9 Q172Y9 A0A1S3DSK7 A0A1D1V639 W5JWC6 A0A1B6LVL5 A0A1B6GVF9 A0A0R3RZ64 A0A182E5H0 A0A2K6VFR7 A0A0B2UU68 W4YQI8 A0A0V0G5Z3 K1R606 A0A0P4VL95 A0A0N5CX61 A0A0P5IGJ1 A0A182TGY3 A0A182MR07 A0A1B6HKV4 A0A0B7BDI0 A0A0N8D1T5 V4B2N8 A0A023EJV1 A0A1S4H1G9 A0A2C9K2T2 A0A3P6UIP3 A0A2S2R0H3 A0A1C7CZP8 A0A182K639 A0A067RCR3 A0A3S1BC59 A0A1I7VXX0 A0A0A9W3J1 A0A182YP94 N6TN55 Q7PVG0 A0A183XCV6 A0A182VL38 A0A0L8GR20 U4UH75 A0A182IJ79 F7ASI0 A0A1S3KBN5 A0A210QQN9 A0A182X0R3 A0A182HGF0 C3YFB1 H2Z1B1 J3JV97 A0A0I9N9F4 A0A182SLZ2 A0A182PG21 A0A0N4TWP8
Ontologies
Topology
Subcellular location
Endoplasmic reticulum membrane
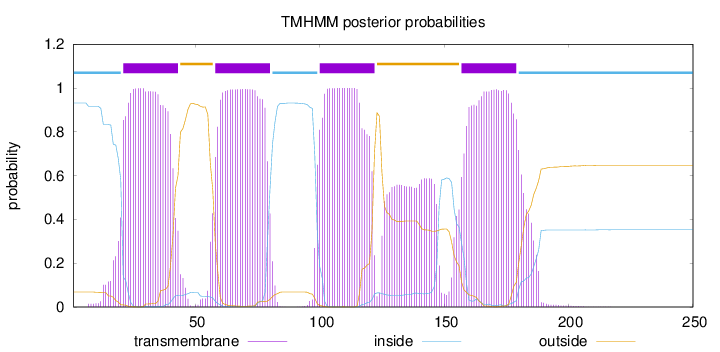
Length:
250
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
105.99449
Exp number, first 60 AAs:
25.82124
Total prob of N-in:
0.93136
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 57
TMhelix
58 - 80
inside
81 - 99
TMhelix
100 - 122
outside
123 - 156
TMhelix
157 - 179
inside
180 - 250
Population Genetic Test Statistics
Pi
228.189427
Theta
184.67821
Tajima's D
0.840377
CLR
0
CSRT
0.607469626518674
Interpretation
Uncertain
