Gene
KWMTBOMO11973
Pre Gene Modal
BGIBMGA004378
Annotation
PREDICTED:_clavesin-2-like_isoform_X1_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 3.164
Sequence
CDS
ATGGCCACTTCAGTCCTCCAACCCGAAGATTGCCGAAGAGAACACGAACCAAAGCCAGACATCATCGAATCACCGCGGCTGACTGATGATGAACGCGTACACATTCTGGAAGCGAAGGAACGAGAGACCGGTGAACTGCCGCCTGAGCTTCGGGAAAAAGCCCGGATCGAACTGAGAGAAGATCCAATTTTGAGAGAAGAAGCTTTGAATCAAATGAGACATTTTATAGAGAAGCATCCTGCGATTAAAAAATGCCGTCAAGATGCACCATTCTTGTTGCGTTTCCTGCGTACAAAGAAATACTCAATACCCCAGGCCTGCTCGATGCTGGAACGGTACCTGACCATACGACAAATGTACCCGCAATGGTTCCAGAAACTGGATCCTCTTGACCCGAAGATAGCTGCAGTTATAGACGCCGGGTATCTAGTGCCATTGCCGAAGAGAGACGCCGAGGGGAGAAGAGTGGTTCTGTCCTGTATGGGACGCTTCGATCCTCACTTATACGACAGTTGTGTAATGGCCCGAGTTCATTCAATGATTGTGGAACACCTCCTCGACGAGCCACGCTCCCAACTTCTCGGCTACACTCATGTTAATGACGAAGCCGGCATGCAAATGCCTCACATTAGTCTTTGGTCTCTCACCGATGTCAGGATAATGCTTAATTGCATTCAGAATTCAACACCGATGAGGCACAAGAGGACACATTTTGTGAACATACCGCATTATGGAGTCAGATTTTTCGAATTTGCAGTTTCGTTGTTGAGTGATAAACTGAAAGACCGTGTTATGTTCCATCGTACGAGTAGTGACCTAACAAAATACGTTGACCCTGCCATTTTGCCAAAAGAATATGGCGGTACAGTCCCACTAAAAGATATGATTGAAGAACTGAAACGTAATTTATTGAAGCAAAGAGAGGAGCTACTGGCCTTGGATGACATGTGCATCGATTTGTATGCACTAGAAAAAAATGATTTGACACAAGACATTCATTCAACCGCCGGATCGTTCCGTAAACTTGAATTGGATTAA
Protein
MATSVLQPEDCRREHEPKPDIIESPRLTDDERVHILEAKERETGELPPELREKARIELREDPILREEALNQMRHFIEKHPAIKKCRQDAPFLLRFLRTKKYSIPQACSMLERYLTIRQMYPQWFQKLDPLDPKIAAVIDAGYLVPLPKRDAEGRRVVLSCMGRFDPHLYDSCVMARVHSMIVEHLLDEPRSQLLGYTHVNDEAGMQMPHISLWSLTDVRIMLNCIQNSTPMRHKRTHFVNIPHYGVRFFEFAVSLLSDKLKDRVMFHRTSSDLTKYVDPAILPKEYGGTVPLKDMIEELKRNLLKQREELLALDDMCIDLYALEKNDLTQDIHSTAGSFRKLELD
Summary
Uniprot
H9J4D9
A0A2A4JFF1
A0A2W1BQD9
I4DJ17
A0A0S2Z351
A0A2H1WNU3
+ More
A0A2H4RMQ0 A0A3S2LDY5 A0A2H4RMT3 A0A212EWJ6 A0A0L7KUX0 B4L080 A0A026X4A6 E2AKP8 A0A0L7RAV0 E9I8F2 A0A1I8P4P8 A0A088AU83 E2BHB2 A0A0N0U5W8 A0A2A3ENT0 A0A1B6FJZ8 B3MB13 K7IMI5 T1PPD2 B4PGR5 A0A1I8N353 A0A1B6JQG9 A0A0C9QEY8 A0A1B0B5J8 A0A310S4U0 A0A1A9Y0T3 A0A1W4UUT4 A0A0M3QWL0 A0A154PP17 A0A1B0AI52 A0A1B0G8G5 A0A1B6LYX5 A0A0L0C613 A0A3B0K409 A0A0Q9WTF0 B4LGN6 A0A195C2P7 W8BNM0 B3NHA8 A0A1A9V3P8 A0A0J7LAZ4 B4HG21 A0A0J9RVF0 Q9VTY1 A0A1J1IDV5 B4GRF4 B5DPD3 B4J322 B4N344 A0A034WGZ8 E0W0G5 A0A3L8DYT5 A0A0A1XFE3 A0A0K8UCP2 A0A195EYP4 F4W5T2 A0A023F2I7 J3JTC8 A0A336M0H0 A0A2P8Z557 A0A139WG06 J3JYV0 A0A2H8TJU0 A0A067QQN5 C4WWS6 J9K7J4 A0A151X2M8 A0A2J7QQQ8 A0A023EPN0 A0A182GWK0 A0A1Q3FMQ4 A0A158P2S1 Q0IET5 A0A2M4BTZ5 B0XBV4 B0X414 A0A2M4BU56 A0A2M3Z4T7 A0A2M3Z4X9 A0A2M3ZH19 A0A195DFG9 A0A1Y1MWL1 W5JFQ5 A0A182IWX1 A0A182NI43 A0A2R7VUT4 A0A182QF89 A0A182FDD8 A0A182PRD7 A0A182M0L7 A0A182I325 A0A182U9N0
A0A2H4RMQ0 A0A3S2LDY5 A0A2H4RMT3 A0A212EWJ6 A0A0L7KUX0 B4L080 A0A026X4A6 E2AKP8 A0A0L7RAV0 E9I8F2 A0A1I8P4P8 A0A088AU83 E2BHB2 A0A0N0U5W8 A0A2A3ENT0 A0A1B6FJZ8 B3MB13 K7IMI5 T1PPD2 B4PGR5 A0A1I8N353 A0A1B6JQG9 A0A0C9QEY8 A0A1B0B5J8 A0A310S4U0 A0A1A9Y0T3 A0A1W4UUT4 A0A0M3QWL0 A0A154PP17 A0A1B0AI52 A0A1B0G8G5 A0A1B6LYX5 A0A0L0C613 A0A3B0K409 A0A0Q9WTF0 B4LGN6 A0A195C2P7 W8BNM0 B3NHA8 A0A1A9V3P8 A0A0J7LAZ4 B4HG21 A0A0J9RVF0 Q9VTY1 A0A1J1IDV5 B4GRF4 B5DPD3 B4J322 B4N344 A0A034WGZ8 E0W0G5 A0A3L8DYT5 A0A0A1XFE3 A0A0K8UCP2 A0A195EYP4 F4W5T2 A0A023F2I7 J3JTC8 A0A336M0H0 A0A2P8Z557 A0A139WG06 J3JYV0 A0A2H8TJU0 A0A067QQN5 C4WWS6 J9K7J4 A0A151X2M8 A0A2J7QQQ8 A0A023EPN0 A0A182GWK0 A0A1Q3FMQ4 A0A158P2S1 Q0IET5 A0A2M4BTZ5 B0XBV4 B0X414 A0A2M4BU56 A0A2M3Z4T7 A0A2M3Z4X9 A0A2M3ZH19 A0A195DFG9 A0A1Y1MWL1 W5JFQ5 A0A182IWX1 A0A182NI43 A0A2R7VUT4 A0A182QF89 A0A182FDD8 A0A182PRD7 A0A182M0L7 A0A182I325 A0A182U9N0
Pubmed
19121390
28756777
22651552
26354079
25684408
29136137
+ More
22118469 26227816 17994087 24508170 20798317 21282665 20075255 17550304 25315136 26108605 24495485 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 25348373 20566863 30249741 25830018 21719571 25474469 22516182 29403074 18362917 19820115 24845553 24945155 26483478 21347285 17510324 28004739 20920257 23761445
22118469 26227816 17994087 24508170 20798317 21282665 20075255 17550304 25315136 26108605 24495485 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 25348373 20566863 30249741 25830018 21719571 25474469 22516182 29403074 18362917 19820115 24845553 24945155 26483478 21347285 17510324 28004739 20920257 23761445
EMBL
BABH01022067
NWSH01001732
PCG70304.1
KZ149954
PZC76511.1
AK401285
+ More
KQ459460 BAM17907.1 KPJ00786.1 KT943543 ALQ33301.1 ODYU01009970 SOQ54743.1 MG434604 ATY51910.1 RSAL01000030 RVE51670.1 MG434645 ATY51951.1 AGBW02011960 OWR45862.1 JTDY01005572 KOB66871.1 CH933809 EDW18026.2 KK107015 EZA62861.1 GL440368 EFN65988.1 KQ414617 KOC68047.1 GL761538 EFZ23179.1 GL448287 EFN84881.1 KQ435752 KOX76208.1 KZ288203 PBC33463.1 GECZ01019268 JAS50501.1 CH902618 EDV40279.2 KA649878 AFP64507.1 CM000159 EDW94304.2 GECU01006286 JAT01421.1 GBYB01001919 JAG71686.1 JXJN01008760 KQ775154 OAD52285.1 CP012525 ALC44333.1 KQ434977 KZC12920.1 CCAG010000253 GEBQ01011084 JAT28893.1 JRES01000951 KNC26854.1 OUUW01000012 SPP87432.1 CH940647 KRF84415.1 EDW69474.2 KQ978379 KYM94463.1 GAMC01003855 JAC02701.1 CH954178 EDV51565.2 LBMM01000070 KMR05077.1 CH480815 EDW41267.1 CM002912 KMY99239.1 AE014296 AY061455 AAF49914.2 AAL29003.1 CVRI01000047 CRK98400.1 CH479188 EDW40339.1 CH379069 EDY73495.2 CH916366 EDV96093.1 CH964062 EDW78783.2 GAKP01004111 JAC54841.1 DS235858 EEB19121.1 QOIP01000002 RLU25630.1 GBXI01004636 JAD09656.1 GDHF01027882 JAI24432.1 KQ981905 KYN33343.1 GL887695 EGI70408.1 GBBI01002885 JAC15827.1 BT126484 AEE61448.1 UFQS01000388 UFQT01000388 SSX03461.1 SSX23826.1 PYGN01000190 PSN51630.1 KQ971351 KYB26819.1 BT128431 AEE63388.1 GFXV01001743 MBW13548.1 KK853083 KDR11656.1 AK342238 BAH72346.1 ABLF02016643 ABLF02035746 KQ982576 KYQ54653.1 NEVH01012083 PNF30920.1 GAPW01002615 JAC10983.1 JXUM01019964 JXUM01094568 KQ564137 KQ560560 KXJ72702.1 KXJ81891.1 GFDL01006184 JAV28861.1 ADTU01007409 ADTU01007410 ADTU01007411 ADTU01007412 ADTU01007413 CH477478 EAT40266.1 GGFJ01007404 MBW56545.1 DS232655 EDS44483.1 DS232327 EDS40091.1 GGFJ01007403 MBW56544.1 GGFM01002775 MBW23526.1 GGFM01002757 MBW23508.1 GGFM01007071 MBW27822.1 KQ980903 KYN11606.1 GEZM01018847 JAV90013.1 ADMH02001280 ETN63207.1 KK854099 PTY11257.1 AXCN02000392 AXCM01002755 APCN01000195
KQ459460 BAM17907.1 KPJ00786.1 KT943543 ALQ33301.1 ODYU01009970 SOQ54743.1 MG434604 ATY51910.1 RSAL01000030 RVE51670.1 MG434645 ATY51951.1 AGBW02011960 OWR45862.1 JTDY01005572 KOB66871.1 CH933809 EDW18026.2 KK107015 EZA62861.1 GL440368 EFN65988.1 KQ414617 KOC68047.1 GL761538 EFZ23179.1 GL448287 EFN84881.1 KQ435752 KOX76208.1 KZ288203 PBC33463.1 GECZ01019268 JAS50501.1 CH902618 EDV40279.2 KA649878 AFP64507.1 CM000159 EDW94304.2 GECU01006286 JAT01421.1 GBYB01001919 JAG71686.1 JXJN01008760 KQ775154 OAD52285.1 CP012525 ALC44333.1 KQ434977 KZC12920.1 CCAG010000253 GEBQ01011084 JAT28893.1 JRES01000951 KNC26854.1 OUUW01000012 SPP87432.1 CH940647 KRF84415.1 EDW69474.2 KQ978379 KYM94463.1 GAMC01003855 JAC02701.1 CH954178 EDV51565.2 LBMM01000070 KMR05077.1 CH480815 EDW41267.1 CM002912 KMY99239.1 AE014296 AY061455 AAF49914.2 AAL29003.1 CVRI01000047 CRK98400.1 CH479188 EDW40339.1 CH379069 EDY73495.2 CH916366 EDV96093.1 CH964062 EDW78783.2 GAKP01004111 JAC54841.1 DS235858 EEB19121.1 QOIP01000002 RLU25630.1 GBXI01004636 JAD09656.1 GDHF01027882 JAI24432.1 KQ981905 KYN33343.1 GL887695 EGI70408.1 GBBI01002885 JAC15827.1 BT126484 AEE61448.1 UFQS01000388 UFQT01000388 SSX03461.1 SSX23826.1 PYGN01000190 PSN51630.1 KQ971351 KYB26819.1 BT128431 AEE63388.1 GFXV01001743 MBW13548.1 KK853083 KDR11656.1 AK342238 BAH72346.1 ABLF02016643 ABLF02035746 KQ982576 KYQ54653.1 NEVH01012083 PNF30920.1 GAPW01002615 JAC10983.1 JXUM01019964 JXUM01094568 KQ564137 KQ560560 KXJ72702.1 KXJ81891.1 GFDL01006184 JAV28861.1 ADTU01007409 ADTU01007410 ADTU01007411 ADTU01007412 ADTU01007413 CH477478 EAT40266.1 GGFJ01007404 MBW56545.1 DS232655 EDS44483.1 DS232327 EDS40091.1 GGFJ01007403 MBW56544.1 GGFM01002775 MBW23526.1 GGFM01002757 MBW23508.1 GGFM01007071 MBW27822.1 KQ980903 KYN11606.1 GEZM01018847 JAV90013.1 ADMH02001280 ETN63207.1 KK854099 PTY11257.1 AXCN02000392 AXCM01002755 APCN01000195
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000007151
UP000037510
+ More
UP000009192 UP000053097 UP000000311 UP000053825 UP000095300 UP000005203 UP000008237 UP000053105 UP000242457 UP000007801 UP000002358 UP000002282 UP000095301 UP000092460 UP000092443 UP000192221 UP000092553 UP000076502 UP000092445 UP000092444 UP000037069 UP000268350 UP000008792 UP000078542 UP000008711 UP000078200 UP000036403 UP000001292 UP000000803 UP000183832 UP000008744 UP000001819 UP000001070 UP000007798 UP000009046 UP000279307 UP000078541 UP000007755 UP000245037 UP000007266 UP000027135 UP000007819 UP000075809 UP000235965 UP000069940 UP000249989 UP000005205 UP000008820 UP000002320 UP000078492 UP000000673 UP000075880 UP000075884 UP000075886 UP000069272 UP000075885 UP000075883 UP000075840 UP000075902
UP000009192 UP000053097 UP000000311 UP000053825 UP000095300 UP000005203 UP000008237 UP000053105 UP000242457 UP000007801 UP000002358 UP000002282 UP000095301 UP000092460 UP000092443 UP000192221 UP000092553 UP000076502 UP000092445 UP000092444 UP000037069 UP000268350 UP000008792 UP000078542 UP000008711 UP000078200 UP000036403 UP000001292 UP000000803 UP000183832 UP000008744 UP000001819 UP000001070 UP000007798 UP000009046 UP000279307 UP000078541 UP000007755 UP000245037 UP000007266 UP000027135 UP000007819 UP000075809 UP000235965 UP000069940 UP000249989 UP000005205 UP000008820 UP000002320 UP000078492 UP000000673 UP000075880 UP000075884 UP000075886 UP000069272 UP000075885 UP000075883 UP000075840 UP000075902
Interpro
Gene 3D
CDD
ProteinModelPortal
H9J4D9
A0A2A4JFF1
A0A2W1BQD9
I4DJ17
A0A0S2Z351
A0A2H1WNU3
+ More
A0A2H4RMQ0 A0A3S2LDY5 A0A2H4RMT3 A0A212EWJ6 A0A0L7KUX0 B4L080 A0A026X4A6 E2AKP8 A0A0L7RAV0 E9I8F2 A0A1I8P4P8 A0A088AU83 E2BHB2 A0A0N0U5W8 A0A2A3ENT0 A0A1B6FJZ8 B3MB13 K7IMI5 T1PPD2 B4PGR5 A0A1I8N353 A0A1B6JQG9 A0A0C9QEY8 A0A1B0B5J8 A0A310S4U0 A0A1A9Y0T3 A0A1W4UUT4 A0A0M3QWL0 A0A154PP17 A0A1B0AI52 A0A1B0G8G5 A0A1B6LYX5 A0A0L0C613 A0A3B0K409 A0A0Q9WTF0 B4LGN6 A0A195C2P7 W8BNM0 B3NHA8 A0A1A9V3P8 A0A0J7LAZ4 B4HG21 A0A0J9RVF0 Q9VTY1 A0A1J1IDV5 B4GRF4 B5DPD3 B4J322 B4N344 A0A034WGZ8 E0W0G5 A0A3L8DYT5 A0A0A1XFE3 A0A0K8UCP2 A0A195EYP4 F4W5T2 A0A023F2I7 J3JTC8 A0A336M0H0 A0A2P8Z557 A0A139WG06 J3JYV0 A0A2H8TJU0 A0A067QQN5 C4WWS6 J9K7J4 A0A151X2M8 A0A2J7QQQ8 A0A023EPN0 A0A182GWK0 A0A1Q3FMQ4 A0A158P2S1 Q0IET5 A0A2M4BTZ5 B0XBV4 B0X414 A0A2M4BU56 A0A2M3Z4T7 A0A2M3Z4X9 A0A2M3ZH19 A0A195DFG9 A0A1Y1MWL1 W5JFQ5 A0A182IWX1 A0A182NI43 A0A2R7VUT4 A0A182QF89 A0A182FDD8 A0A182PRD7 A0A182M0L7 A0A182I325 A0A182U9N0
A0A2H4RMQ0 A0A3S2LDY5 A0A2H4RMT3 A0A212EWJ6 A0A0L7KUX0 B4L080 A0A026X4A6 E2AKP8 A0A0L7RAV0 E9I8F2 A0A1I8P4P8 A0A088AU83 E2BHB2 A0A0N0U5W8 A0A2A3ENT0 A0A1B6FJZ8 B3MB13 K7IMI5 T1PPD2 B4PGR5 A0A1I8N353 A0A1B6JQG9 A0A0C9QEY8 A0A1B0B5J8 A0A310S4U0 A0A1A9Y0T3 A0A1W4UUT4 A0A0M3QWL0 A0A154PP17 A0A1B0AI52 A0A1B0G8G5 A0A1B6LYX5 A0A0L0C613 A0A3B0K409 A0A0Q9WTF0 B4LGN6 A0A195C2P7 W8BNM0 B3NHA8 A0A1A9V3P8 A0A0J7LAZ4 B4HG21 A0A0J9RVF0 Q9VTY1 A0A1J1IDV5 B4GRF4 B5DPD3 B4J322 B4N344 A0A034WGZ8 E0W0G5 A0A3L8DYT5 A0A0A1XFE3 A0A0K8UCP2 A0A195EYP4 F4W5T2 A0A023F2I7 J3JTC8 A0A336M0H0 A0A2P8Z557 A0A139WG06 J3JYV0 A0A2H8TJU0 A0A067QQN5 C4WWS6 J9K7J4 A0A151X2M8 A0A2J7QQQ8 A0A023EPN0 A0A182GWK0 A0A1Q3FMQ4 A0A158P2S1 Q0IET5 A0A2M4BTZ5 B0XBV4 B0X414 A0A2M4BU56 A0A2M3Z4T7 A0A2M3Z4X9 A0A2M3ZH19 A0A195DFG9 A0A1Y1MWL1 W5JFQ5 A0A182IWX1 A0A182NI43 A0A2R7VUT4 A0A182QF89 A0A182FDD8 A0A182PRD7 A0A182M0L7 A0A182I325 A0A182U9N0
PDB
3HY5
E-value=4.54837e-26,
Score=292
Ontologies
GO
Topology
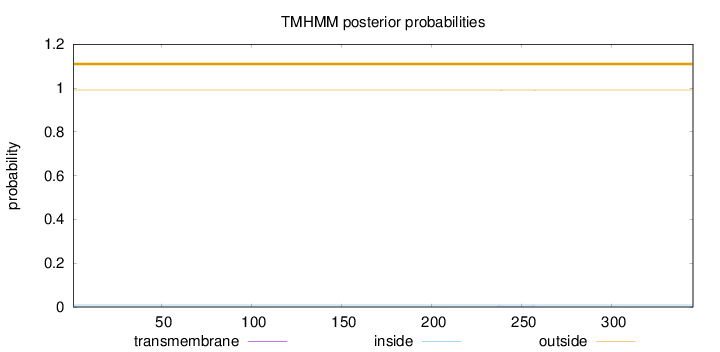
Length:
345
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01596
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00891
outside
1 - 345
Population Genetic Test Statistics
Pi
212.622019
Theta
169.756576
Tajima's D
0.998437
CLR
0.278672
CSRT
0.653867306634668
Interpretation
Uncertain
