Gene
KWMTBOMO11964
Pre Gene Modal
BGIBMGA004481
Annotation
PREDICTED:_uncharacterized_protein_LOC101744636_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.193
Sequence
CDS
ATGTCAAACTTTGTTGACAGAGCGTCTGACCGATCTCCGTTCGATGGTTCTTGCTGTTCCGGATACAGACCTGTTGATTCTGCGAGCCTCACCACGCCCGATGAAGACGAAATGGACCCAATTCCGAAGTATAGAGTGGTGTTGCTCGGCGATGCGGGAGTCGGAAAAACAGCCCTGGTATCACAATTTATGACGTCAGAATACATGAATACATACGACGCTAGTTTAGACGACGAATTCGGTGAGAAGTCTGTCTCGGTGTTGCTCGACGGAGAAGAATCAGAGCTCATCTTCATCGATCATCCCAGCACTGAGATGTCCATCGAGAACTGTTTGTCGACGTATGAGCCACATGCGTGCGTGGTTGTTTACTCTGTGGTCGCCAAGAGCTCGCTGCAAAGGGCCTCGGATCTGTTAGCTTACCTGTGCAGAGAACAATTCACGGTCGACAGAACTGTTGTGCTGGTCGGGAACAAGGCGGACTTGGCGAGGGCCAGACAAGTCTCGACTAATGAAGGCAAAGCACTCGCAACGTCCAGAGACTGTAAGTTCATCGAGACATCCTCCGGAATCCAGCACAATGTTGACGAACTGCTCGTCGGTATACTGAAGCAGATACGTTTGAAAGAAACAAGAGACAAGAAGCAAGCCAAGAAAACTTCAACTACCAAGGAAGCCAAACCAACAAAACTAGCCAGCTCTCGCACTCATATCTCGCTGAGCATCGCAAGGGAACTGCTACAGAAAATATGCATCAACGATATATCGAAATCAAAGTCCTGCGAGAATCTACACGTTCTCTAA
Protein
MSNFVDRASDRSPFDGSCCSGYRPVDSASLTTPDEDEMDPIPKYRVVLLGDAGVGKTALVSQFMTSEYMNTYDASLDDEFGEKSVSVLLDGEESELIFIDHPSTEMSIENCLSTYEPHACVVVYSVVAKSSLQRASDLLAYLCREQFTVDRTVVLVGNKADLARARQVSTNEGKALATSRDCKFIETSSGIQHNVDELLVGILKQIRLKETRDKKQAKKTSTTKEAKPTKLASSRTHISLSIARELLQKICINDISKSKSCENLHVL
Summary
Uniprot
A0A2A4JPS3
A0A194Q5Q7
A0A2W1BXK6
A0A0N0PEB3
A0A1W4WK65
H9J4P1
+ More
N6TPT7 A0A067R586 A0A1A9VXH3 A0A1J1HY87 A0A232FMR0 A0A076N3M5 E2BGK3 A0A154PSA7 A0A1B0A7D9 B3NMU5 Q6NN22 W8C1L6 A0A0J9RFY7 A0A1W4VDY5 B4P4C9 A0A0A1X9I6 A0A0M4EU59 A0A0C9RF24 B4KTA8 A0A1A9Y431 B4LJG0 A0A2H1W924 A0A034WM71 A0A1B0BI46 B3MDF7 B4JW29 A0A3B0J1W7 A0A0B4LFV4 A0A0J9RFR4 A0A1W4VRE5 A0A0R1DYR0 K7J0I3 B4GGR9 B4N5X3 A0A0R3NMY1 A0A0Q9XA36 A0A1B0G235 A0A182MMW0 A0A182UWR2 A0A182X6C5 Q7QJQ5 A0A182TQY6 A0A182I3N1 A0A182WIQ1 A0A0Q9W319 A0A182P3L6 A0A182FFV4 A0A182KRS2 A0A182QUK6 Q28ZU5 Q17KU9 B0VZD4 A0A0R3NMY9 U4U8U3 A0A1B0C919 W5JF38 A0A182G815 A0A087ZYN3 A0A1Y1MUT9 A0A084VWZ6 A0A2S2R2Y9 A0A0L7RE47 E0VTM1 A0A2H8TL51 A0A336MN11 A0A1I8PL73 B4QCZ0 A0A2S2NCL8 A0A182XZ77 D6WSB6 J9JRB0 A0A026WYH1 A0A1A9W9T6 A0A3L8DRB9 E2B0R8 A0A0P6BD89 A0A1I8M6U6 E9GZQ3 A0A0P6IM08 A0A182RFL9 A0A310ST19 A0A182NHN0 A0A2R7X3T4 E0W0D5 A0A0N0BKR8 V9IAD2 A0A067RH09 V9IAG9 A0A0L7RB28 A0A026WPL9 A0A3L8DYI7 A0A158P2S2 E2BHB0 F4W5T4
N6TPT7 A0A067R586 A0A1A9VXH3 A0A1J1HY87 A0A232FMR0 A0A076N3M5 E2BGK3 A0A154PSA7 A0A1B0A7D9 B3NMU5 Q6NN22 W8C1L6 A0A0J9RFY7 A0A1W4VDY5 B4P4C9 A0A0A1X9I6 A0A0M4EU59 A0A0C9RF24 B4KTA8 A0A1A9Y431 B4LJG0 A0A2H1W924 A0A034WM71 A0A1B0BI46 B3MDF7 B4JW29 A0A3B0J1W7 A0A0B4LFV4 A0A0J9RFR4 A0A1W4VRE5 A0A0R1DYR0 K7J0I3 B4GGR9 B4N5X3 A0A0R3NMY1 A0A0Q9XA36 A0A1B0G235 A0A182MMW0 A0A182UWR2 A0A182X6C5 Q7QJQ5 A0A182TQY6 A0A182I3N1 A0A182WIQ1 A0A0Q9W319 A0A182P3L6 A0A182FFV4 A0A182KRS2 A0A182QUK6 Q28ZU5 Q17KU9 B0VZD4 A0A0R3NMY9 U4U8U3 A0A1B0C919 W5JF38 A0A182G815 A0A087ZYN3 A0A1Y1MUT9 A0A084VWZ6 A0A2S2R2Y9 A0A0L7RE47 E0VTM1 A0A2H8TL51 A0A336MN11 A0A1I8PL73 B4QCZ0 A0A2S2NCL8 A0A182XZ77 D6WSB6 J9JRB0 A0A026WYH1 A0A1A9W9T6 A0A3L8DRB9 E2B0R8 A0A0P6BD89 A0A1I8M6U6 E9GZQ3 A0A0P6IM08 A0A182RFL9 A0A310ST19 A0A182NHN0 A0A2R7X3T4 E0W0D5 A0A0N0BKR8 V9IAD2 A0A067RH09 V9IAG9 A0A0L7RB28 A0A026WPL9 A0A3L8DYI7 A0A158P2S2 E2BHB0 F4W5T4
Pubmed
26354079
28756777
19121390
23537049
24845553
28648823
+ More
20798317 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 22936249 17550304 25830018 25348373 20075255 15632085 18057021 12364791 14747013 17210077 20966253 23185243 17510324 20920257 23761445 26483478 28004739 24438588 20566863 25244985 18362917 19820115 24508170 30249741 25315136 21292972 21347285 21719571
20798317 17994087 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 24495485 22936249 17550304 25830018 25348373 20075255 15632085 18057021 12364791 14747013 17210077 20966253 23185243 17510324 20920257 23761445 26483478 28004739 24438588 20566863 25244985 18362917 19820115 24508170 30249741 25315136 21292972 21347285 21719571
EMBL
NWSH01000940
PCG73443.1
KQ459460
KPJ00689.1
KZ149954
PZC76503.1
+ More
KQ460009 KPJ18476.1 BABH01022037 BABH01022038 APGK01025866 KB740605 ENN80018.1 KK852686 KDR18450.1 CVRI01000030 CRK92522.1 NNAY01000030 OXU31799.1 KJ826442 AIJ28976.1 GL448189 EFN85196.1 KQ435037 KZC14148.1 CH954179 EDV55099.1 AE013599 BT011477 AAF57675.3 AAR99135.1 GAMC01011002 GAMC01011001 JAB95554.1 CM002911 KMY94877.1 CM000158 EDW91615.1 GBXI01006907 JAD07385.1 CP012524 ALC41175.1 GBYB01006990 JAG76757.1 CH933808 EDW10620.1 CH940648 EDW60540.1 ODYU01006993 SOQ49342.1 GAKP01003228 JAC55724.1 JXJN01014771 CH902619 EDV36405.1 CH916375 EDV98167.1 OUUW01000001 SPP75027.1 AHN56372.1 KMY94875.1 KMY94876.1 KMY94878.1 KRJ99990.1 KRJ99991.1 CH479183 EDW35689.1 CH964154 EDW79762.1 CM000071 KRT02282.1 KRG05355.1 KRG05356.1 CCAG010020580 AXCM01000215 AAAB01008807 EAA04508.3 APCN01000215 KRF79466.1 KRF79467.1 KRF79468.1 AXCN02000593 EAL25517.3 KRT02283.1 CH477221 EAT47305.1 DS231814 EDS31671.1 KRT02281.1 KB632095 ERL88778.1 AJWK01001779 AJWK01001780 ADMH02001692 ETN61414.1 JXUM01007950 JXUM01007951 JXUM01007952 KQ560250 KXJ83564.1 GEZM01020311 JAV89309.1 ATLV01017810 KE525192 KFB42490.1 GGMS01015156 MBY84359.1 KQ414613 KOC69090.1 DS235767 EEB16696.1 GFXV01002925 MBW14730.1 UFQS01001592 UFQS01001954 UFQT01001592 UFQT01001954 SSX11528.1 SSX12779.1 SSX31095.1 CM000362 EDX07695.1 GGMR01002239 MBY14858.1 KQ971352 EFA06392.2 ABLF02034772 ABLF02034773 ABLF02034774 ABLF02034775 ABLF02034776 ABLF02034777 ABLF02034779 ABLF02034781 ABLF02034782 ABLF02043290 KK107063 EZA61082.1 QOIP01000005 RLU22960.1 GL444666 EFN60740.1 GDIP01017908 LRGB01003380 JAM85807.1 KZS02896.1 GL732578 EFX75065.1 GDIQ01002927 JAN91810.1 KQ760243 OAD61265.1 KK856832 PTY26459.1 DS235858 EEB19091.1 KQ435694 KOX80984.1 JR037995 AEY58068.1 KDR18448.1 JR037997 AEY58070.1 KQ414617 KOC68030.1 KK107138 EZA57923.1 QOIP01000002 RLU25497.1 ADTU01007418 ADTU01007419 ADTU01007420 ADTU01007421 ADTU01007422 GL448287 EFN84879.1 GL887695 EGI70410.1
KQ460009 KPJ18476.1 BABH01022037 BABH01022038 APGK01025866 KB740605 ENN80018.1 KK852686 KDR18450.1 CVRI01000030 CRK92522.1 NNAY01000030 OXU31799.1 KJ826442 AIJ28976.1 GL448189 EFN85196.1 KQ435037 KZC14148.1 CH954179 EDV55099.1 AE013599 BT011477 AAF57675.3 AAR99135.1 GAMC01011002 GAMC01011001 JAB95554.1 CM002911 KMY94877.1 CM000158 EDW91615.1 GBXI01006907 JAD07385.1 CP012524 ALC41175.1 GBYB01006990 JAG76757.1 CH933808 EDW10620.1 CH940648 EDW60540.1 ODYU01006993 SOQ49342.1 GAKP01003228 JAC55724.1 JXJN01014771 CH902619 EDV36405.1 CH916375 EDV98167.1 OUUW01000001 SPP75027.1 AHN56372.1 KMY94875.1 KMY94876.1 KMY94878.1 KRJ99990.1 KRJ99991.1 CH479183 EDW35689.1 CH964154 EDW79762.1 CM000071 KRT02282.1 KRG05355.1 KRG05356.1 CCAG010020580 AXCM01000215 AAAB01008807 EAA04508.3 APCN01000215 KRF79466.1 KRF79467.1 KRF79468.1 AXCN02000593 EAL25517.3 KRT02283.1 CH477221 EAT47305.1 DS231814 EDS31671.1 KRT02281.1 KB632095 ERL88778.1 AJWK01001779 AJWK01001780 ADMH02001692 ETN61414.1 JXUM01007950 JXUM01007951 JXUM01007952 KQ560250 KXJ83564.1 GEZM01020311 JAV89309.1 ATLV01017810 KE525192 KFB42490.1 GGMS01015156 MBY84359.1 KQ414613 KOC69090.1 DS235767 EEB16696.1 GFXV01002925 MBW14730.1 UFQS01001592 UFQS01001954 UFQT01001592 UFQT01001954 SSX11528.1 SSX12779.1 SSX31095.1 CM000362 EDX07695.1 GGMR01002239 MBY14858.1 KQ971352 EFA06392.2 ABLF02034772 ABLF02034773 ABLF02034774 ABLF02034775 ABLF02034776 ABLF02034777 ABLF02034779 ABLF02034781 ABLF02034782 ABLF02043290 KK107063 EZA61082.1 QOIP01000005 RLU22960.1 GL444666 EFN60740.1 GDIP01017908 LRGB01003380 JAM85807.1 KZS02896.1 GL732578 EFX75065.1 GDIQ01002927 JAN91810.1 KQ760243 OAD61265.1 KK856832 PTY26459.1 DS235858 EEB19091.1 KQ435694 KOX80984.1 JR037995 AEY58068.1 KDR18448.1 JR037997 AEY58070.1 KQ414617 KOC68030.1 KK107138 EZA57923.1 QOIP01000002 RLU25497.1 ADTU01007418 ADTU01007419 ADTU01007420 ADTU01007421 ADTU01007422 GL448287 EFN84879.1 GL887695 EGI70410.1
Proteomes
UP000218220
UP000053268
UP000053240
UP000192223
UP000005204
UP000019118
+ More
UP000027135 UP000078200 UP000183832 UP000215335 UP000008237 UP000076502 UP000092445 UP000008711 UP000000803 UP000192221 UP000002282 UP000092553 UP000009192 UP000092443 UP000008792 UP000092460 UP000007801 UP000001070 UP000268350 UP000002358 UP000008744 UP000007798 UP000001819 UP000092444 UP000075883 UP000075903 UP000076407 UP000007062 UP000075902 UP000075840 UP000075920 UP000075885 UP000069272 UP000075882 UP000075886 UP000008820 UP000002320 UP000030742 UP000092461 UP000000673 UP000069940 UP000249989 UP000005203 UP000030765 UP000053825 UP000009046 UP000095300 UP000000304 UP000076408 UP000007266 UP000007819 UP000053097 UP000091820 UP000279307 UP000000311 UP000076858 UP000095301 UP000000305 UP000075900 UP000075884 UP000053105 UP000005205 UP000007755
UP000027135 UP000078200 UP000183832 UP000215335 UP000008237 UP000076502 UP000092445 UP000008711 UP000000803 UP000192221 UP000002282 UP000092553 UP000009192 UP000092443 UP000008792 UP000092460 UP000007801 UP000001070 UP000268350 UP000002358 UP000008744 UP000007798 UP000001819 UP000092444 UP000075883 UP000075903 UP000076407 UP000007062 UP000075902 UP000075840 UP000075920 UP000075885 UP000069272 UP000075882 UP000075886 UP000008820 UP000002320 UP000030742 UP000092461 UP000000673 UP000069940 UP000249989 UP000005203 UP000030765 UP000053825 UP000009046 UP000095300 UP000000304 UP000076408 UP000007266 UP000007819 UP000053097 UP000091820 UP000279307 UP000000311 UP000076858 UP000095301 UP000000305 UP000075900 UP000075884 UP000053105 UP000005205 UP000007755
Pfam
PF00071 Ras
Interpro
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A2A4JPS3
A0A194Q5Q7
A0A2W1BXK6
A0A0N0PEB3
A0A1W4WK65
H9J4P1
+ More
N6TPT7 A0A067R586 A0A1A9VXH3 A0A1J1HY87 A0A232FMR0 A0A076N3M5 E2BGK3 A0A154PSA7 A0A1B0A7D9 B3NMU5 Q6NN22 W8C1L6 A0A0J9RFY7 A0A1W4VDY5 B4P4C9 A0A0A1X9I6 A0A0M4EU59 A0A0C9RF24 B4KTA8 A0A1A9Y431 B4LJG0 A0A2H1W924 A0A034WM71 A0A1B0BI46 B3MDF7 B4JW29 A0A3B0J1W7 A0A0B4LFV4 A0A0J9RFR4 A0A1W4VRE5 A0A0R1DYR0 K7J0I3 B4GGR9 B4N5X3 A0A0R3NMY1 A0A0Q9XA36 A0A1B0G235 A0A182MMW0 A0A182UWR2 A0A182X6C5 Q7QJQ5 A0A182TQY6 A0A182I3N1 A0A182WIQ1 A0A0Q9W319 A0A182P3L6 A0A182FFV4 A0A182KRS2 A0A182QUK6 Q28ZU5 Q17KU9 B0VZD4 A0A0R3NMY9 U4U8U3 A0A1B0C919 W5JF38 A0A182G815 A0A087ZYN3 A0A1Y1MUT9 A0A084VWZ6 A0A2S2R2Y9 A0A0L7RE47 E0VTM1 A0A2H8TL51 A0A336MN11 A0A1I8PL73 B4QCZ0 A0A2S2NCL8 A0A182XZ77 D6WSB6 J9JRB0 A0A026WYH1 A0A1A9W9T6 A0A3L8DRB9 E2B0R8 A0A0P6BD89 A0A1I8M6U6 E9GZQ3 A0A0P6IM08 A0A182RFL9 A0A310ST19 A0A182NHN0 A0A2R7X3T4 E0W0D5 A0A0N0BKR8 V9IAD2 A0A067RH09 V9IAG9 A0A0L7RB28 A0A026WPL9 A0A3L8DYI7 A0A158P2S2 E2BHB0 F4W5T4
N6TPT7 A0A067R586 A0A1A9VXH3 A0A1J1HY87 A0A232FMR0 A0A076N3M5 E2BGK3 A0A154PSA7 A0A1B0A7D9 B3NMU5 Q6NN22 W8C1L6 A0A0J9RFY7 A0A1W4VDY5 B4P4C9 A0A0A1X9I6 A0A0M4EU59 A0A0C9RF24 B4KTA8 A0A1A9Y431 B4LJG0 A0A2H1W924 A0A034WM71 A0A1B0BI46 B3MDF7 B4JW29 A0A3B0J1W7 A0A0B4LFV4 A0A0J9RFR4 A0A1W4VRE5 A0A0R1DYR0 K7J0I3 B4GGR9 B4N5X3 A0A0R3NMY1 A0A0Q9XA36 A0A1B0G235 A0A182MMW0 A0A182UWR2 A0A182X6C5 Q7QJQ5 A0A182TQY6 A0A182I3N1 A0A182WIQ1 A0A0Q9W319 A0A182P3L6 A0A182FFV4 A0A182KRS2 A0A182QUK6 Q28ZU5 Q17KU9 B0VZD4 A0A0R3NMY9 U4U8U3 A0A1B0C919 W5JF38 A0A182G815 A0A087ZYN3 A0A1Y1MUT9 A0A084VWZ6 A0A2S2R2Y9 A0A0L7RE47 E0VTM1 A0A2H8TL51 A0A336MN11 A0A1I8PL73 B4QCZ0 A0A2S2NCL8 A0A182XZ77 D6WSB6 J9JRB0 A0A026WYH1 A0A1A9W9T6 A0A3L8DRB9 E2B0R8 A0A0P6BD89 A0A1I8M6U6 E9GZQ3 A0A0P6IM08 A0A182RFL9 A0A310ST19 A0A182NHN0 A0A2R7X3T4 E0W0D5 A0A0N0BKR8 V9IAD2 A0A067RH09 V9IAG9 A0A0L7RB28 A0A026WPL9 A0A3L8DYI7 A0A158P2S2 E2BHB0 F4W5T4
PDB
2CJW
E-value=8.26855e-27,
Score=297
Ontologies
KEGG
GO
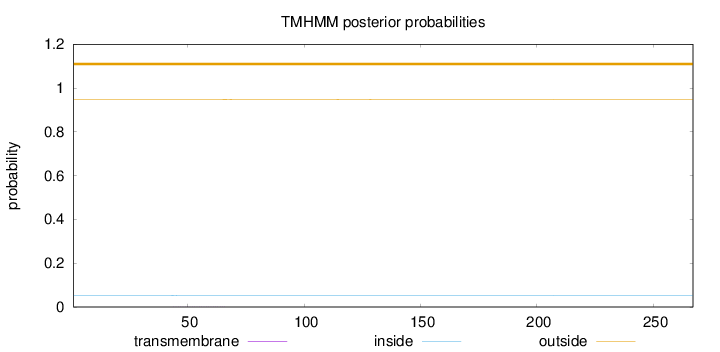
Topology
Length:
267
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02055
Exp number, first 60 AAs:
0.01543
Total prob of N-in:
0.05204
outside
1 - 267
Population Genetic Test Statistics
Pi
186.174955
Theta
157.291797
Tajima's D
0.726926
CLR
0.109778
CSRT
0.581620918954052
Interpretation
Uncertain
