Gene
KWMTBOMO11963
Annotation
PREDICTED:_uncharacterized_protein_LOC101735858_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.612
Sequence
CDS
ATGGTTGCTACTACTGGGGAACTGATACGTGGTAAGTTTAATCTTGAAAATCGCGCTACTTTTAACGTTTGTCATGTAAACGCGCAAAGTATAGCTAGTCACTACAGTGAACTAGATGTAACTTTCACCGACGCCAACGTGCACGCCGTCCTTGTTTCTGAGAGCTGGCTCAAACCTCATCTTCCTTCCGCCGCATACCCCCTACCTGGATTTATTCTAATCCGAAATGATCGGGTGGACAGGAGAGGGGGTGGTGTGGCTATCTATCTCCAAGCCTCCTTTTCTTATAAAGTCATAGCCTCTTCCTCTTCCACGAGTTCCAATTCCGCTGAATACCTTTTCCTTGAGGTTTGGGTCAAGGGAGTCAAAACTTTACTTGGTGTGGTTTATTGTCCTCCCTCTGTTGATTACTTCTCCAGTCTTGACTCAACTCTGGAATCTTTAGGGTCAGATTACGCACACCACATCATCATGGGAGACTTCAACACCGACCTCCTTGTCCCTTTGTCCACTCGTTCACGAAAACTCCTTGACCTTGTTGAGTCAGTCAGTTTACACATCCTCCCTTTACAAGCCACTCACCACAACATTGAAGGAGAGGATACCTGGCTTGACCTAATACTCACTTCCACTCCCGATCTTGTTCATTCTCACGGTCAGTTTCCTGCTCCTGGTTTCTCCCATCATGACCTCATTTACTTATCCTACGTCCTTAAACCTCCCAAATCCAAGCCTAAGGTTTTGCGCGTGCGTTGTTTTGGTCGCATGGATGGACGCAAGTTGTGTGAGGATGCTGCTAAAATTACTTGGGATGAACTGACGGCAGCTACATCTATTGACGAGAAGGTGACTATATTTAACGCTCAACTCCAGGCACTTTATGACACTCATGCTCCCGTCAGGAAATTCAAGTTGAAACGTCCTCCAGCACCTTGGATGACGAGTGAGGTTCGGATGGCGATGAAGCGTAGGGATCGGGCCTTGGCAAAATTCAGGAGACGCCGCACGGAGGAAAACTGGACATCATTTAAGGTTGCGAGGAATTGGTGTAATCAGATGGTACGGAACGCTAAGCGCCGCCATATTCTCGAAAATATAACCTCCGCTTCTTCAGCTAATATCTGGAAGTTCCTTAAGACTCTGGGTATTGGTAGAGAGAAGAATAAAGACTTCCAGGGGACCGTTGCCTTAGATGACCTTAACAGGCATTTCAGCGCATCTGCCGGCATTGATCATCAAGTCAAAAGTCTCACTATTGAATTTATAGCTGGGTTACCTAAACCAGACATTGACCCCTTCCAATTTTCTCCAATGACTTCAGATGATATTAAAAAGATTATCCTATCCATTAGATCTAATGCTGTTGGTTATGACAAAATCACCCGTCGCATGATTACCACAGCTTTAGACCATCTTATTCTCACCATAACCCACATCATTAATTTCTCCTTTAGTACCGACATGTTTCCCTCCCAATGGCGTAAGGCTTATGTCATTCCTCTTCCTAAAATACCGAATCCTACCCTTGTTACTCACTTCCGACCTATATCCATACTTCCTTTCCTATCTAAGGTTTTAGAATCTTGTGCGCACAAACAACTGTCTAACTTTATTCATTCACACAATTTATTGAGTACACTGCAGTCCGGTTTCAGACCTTGCCACAGCACTACTACCGCACTCCTTAAAGTGATTGGCGATATTAGAGCAGGTATGGAGGACACAAAGGTTACGGTATTGGTTCTGATAGACTTCTCAAACGCTTTTAATAACGTCAATCACGACATCCTCTTATCCATTCTCTCCCATCTTGGGATGTCATCTGGAGCATTGGATTGGTTCTCGTCCTATCTTCGAGGTCGTCAGCAGTCGATACACATCGGCGAATCTTCGTCTAGTTGCTTTATGCTCAGGCTGGAGTCGATAGTGTGTCTTCGGCAGTTGGAAAAATAA
Protein
MVATTGELIRGKFNLENRATFNVCHVNAQSIASHYSELDVTFTDANVHAVLVSESWLKPHLPSAAYPLPGFILIRNDRVDRRGGGVAIYLQASFSYKVIASSSSTSSNSAEYLFLEVWVKGVKTLLGVVYCPPSVDYFSSLDSTLESLGSDYAHHIIMGDFNTDLLVPLSTRSRKLLDLVESVSLHILPLQATHHNIEGEDTWLDLILTSTPDLVHSHGQFPAPGFSHHDLIYLSYVLKPPKSKPKVLRVRCFGRMDGRKLCEDAAKITWDELTAATSIDEKVTIFNAQLQALYDTHAPVRKFKLKRPPAPWMTSEVRMAMKRRDRALAKFRRRRTEENWTSFKVARNWCNQMVRNAKRRHILENITSASSANIWKFLKTLGIGREKNKDFQGTVALDDLNRHFSASAGIDHQVKSLTIEFIAGLPKPDIDPFQFSPMTSDDIKKIILSIRSNAVGYDKITRRMITTALDHLILTITHIINFSFSTDMFPSQWRKAYVIPLPKIPNPTLVTHFRPISILPFLSKVLESCAHKQLSNFIHSHNLLSTLQSGFRPCHSTTTALLKVIGDIRAGMEDTKVTVLVLIDFSNAFNNVNHDILLSILSHLGMSSGALDWFSSYLRGRQQSIHIGESSSSCFMLRLESIVCLRQLEK
Summary
Uniprot
A0A2A4JRM8
A0A2A4JPY6
A0A3S2P9A9
A0A3S2TB34
A0A1B6L6Y3
A0A0J7K589
+ More
A0A0J7N335 A0A0A9YIX2 A0A354GJG9 A0A1B6KD49 A0A3C1S1N3 A0A3C1S2P1 A0A1S3DM39 U5ENG8 A0A1W7R6E4 A0A1W7R6F6 A0A3L8D9C4 A0A3L8DQD7 A0A1W7R6K8 U5EQV9 A0A1W7R6J3 A0A3C1RY27 A0A3C1RYE8 A0A3L8DSB3 A0A354GES4 A0A3C1S360 A0A1B6MC85 A0A354GG23 A0A0A9Z9S6 A0A1Y1M3U5 U5EEL4 W8B6W6 A0A2B4RHP5 A0A354GHC6 A0A1Y1LSZ2 A0A354GI57 A0A1W7R628 U5EVC6 A0A2B4R4J4 A0A2B4SCV9 A0A354GIY0 A0A3B3BNA3 W8AIM4 A0A1Y1L0L4 A0A1Y1LAN8 A0A1Y1LAP1 A0A3P9M960 A0A2A4JF59 A0A3B3HIF3 A0A2A4KAV4 A0A3B3HCE9 U5EQB9 A0A3B3HNZ2 A0A1Y1KZ78 A0A0J7NDM1 A0A3P8PBK6 A0A3P9IXQ6 A0A3B3DM95 W4Z8W8 K7JAP1 A0A3B1JMQ8 K7JTK2 A0A3P9K1B3 A0A1Y1LFG1 A0A1Y1LAM2 U5ETR7 A0A2W1BK42 A0A2B4SVL4 A0A2A4IWQ4 A0A3C1RZS6 A0A182GI84 A0A1Y1N9S9
A0A0J7N335 A0A0A9YIX2 A0A354GJG9 A0A1B6KD49 A0A3C1S1N3 A0A3C1S2P1 A0A1S3DM39 U5ENG8 A0A1W7R6E4 A0A1W7R6F6 A0A3L8D9C4 A0A3L8DQD7 A0A1W7R6K8 U5EQV9 A0A1W7R6J3 A0A3C1RY27 A0A3C1RYE8 A0A3L8DSB3 A0A354GES4 A0A3C1S360 A0A1B6MC85 A0A354GG23 A0A0A9Z9S6 A0A1Y1M3U5 U5EEL4 W8B6W6 A0A2B4RHP5 A0A354GHC6 A0A1Y1LSZ2 A0A354GI57 A0A1W7R628 U5EVC6 A0A2B4R4J4 A0A2B4SCV9 A0A354GIY0 A0A3B3BNA3 W8AIM4 A0A1Y1L0L4 A0A1Y1LAN8 A0A1Y1LAP1 A0A3P9M960 A0A2A4JF59 A0A3B3HIF3 A0A2A4KAV4 A0A3B3HCE9 U5EQB9 A0A3B3HNZ2 A0A1Y1KZ78 A0A0J7NDM1 A0A3P8PBK6 A0A3P9IXQ6 A0A3B3DM95 W4Z8W8 K7JAP1 A0A3B1JMQ8 K7JTK2 A0A3P9K1B3 A0A1Y1LFG1 A0A1Y1LAM2 U5ETR7 A0A2W1BK42 A0A2B4SVL4 A0A2A4IWQ4 A0A3C1RZS6 A0A182GI84 A0A1Y1N9S9
Pubmed
EMBL
NWSH01000817
PCG74070.1
NWSH01000893
PCG73654.1
RSAL01000177
RVE45007.1
+ More
RSAL01001570 RVE40758.1 GEBQ01020572 JAT19405.1 LBMM01014048 KMQ85356.1 LBMM01010997 KMQ87090.1 GBHO01012041 GBHO01012038 JAG31563.1 JAG31566.1 DNUT01000507 HBI41157.1 GEBQ01030598 JAT09379.1 DMQF01000352 HAO15199.1 DMQF01000361 HAO15230.1 GANO01004094 JAB55777.1 GEHC01000909 JAV46736.1 GEHC01000896 JAV46749.1 QOIP01000011 RLU16479.1 QOIP01000005 RLU22413.1 GEHC01000940 JAV46705.1 GANO01004066 JAB55805.1 GEHC01000892 JAV46753.1 DMQF01000003 HAO13943.1 DMQF01000030 HAO14052.1 QOIP01000004 RLU23301.1 DNUT01000056 HBI39512.1 DMQF01000478 HAO15711.1 GEBQ01006447 JAT33530.1 DNUT01000170 HBI39961.1 GBHO01002973 JAG40631.1 GEZM01041318 GEZM01041317 JAV80524.1 GANO01004275 JAB55596.1 GAMC01021037 GAMC01021036 JAB85519.1 LSMT01000554 PFX16333.1 DNUT01000297 HBI40414.1 GEZM01051381 GEZM01051380 JAV74995.1 DNUT01000374 HBI40695.1 GEHC01001026 JAV46619.1 GANO01001936 JAB57935.1 LSMT01001965 PFX11729.1 LSMT01000121 PFX26660.1 DNUT01000451 HBI40968.1 GAMC01021038 GAMC01021033 GAMC01021031 JAB85522.1 GEZM01074146 JAV64657.1 GEZM01061184 JAV70703.1 GEZM01061183 JAV70704.1 NWSH01001817 PCG70032.1 NWSH01000010 PCG80923.1 GANO01004361 JAB55510.1 GEZM01074147 JAV64656.1 LBMM01006440 KMQ90605.1 AAGJ04020328 AAZX01023386 AAZX01004038 GEZM01061182 JAV70705.1 GEZM01061185 JAV70702.1 GANO01004238 JAB55633.1 KZ150070 PZC74075.1 LSMT01000021 PFX32582.1 NWSH01006271 PCG63543.1 DMQF01000157 HAO14486.1 JXUM01065503 KQ562350 KXJ76100.1 GEZM01008907 JAV94684.1
RSAL01001570 RVE40758.1 GEBQ01020572 JAT19405.1 LBMM01014048 KMQ85356.1 LBMM01010997 KMQ87090.1 GBHO01012041 GBHO01012038 JAG31563.1 JAG31566.1 DNUT01000507 HBI41157.1 GEBQ01030598 JAT09379.1 DMQF01000352 HAO15199.1 DMQF01000361 HAO15230.1 GANO01004094 JAB55777.1 GEHC01000909 JAV46736.1 GEHC01000896 JAV46749.1 QOIP01000011 RLU16479.1 QOIP01000005 RLU22413.1 GEHC01000940 JAV46705.1 GANO01004066 JAB55805.1 GEHC01000892 JAV46753.1 DMQF01000003 HAO13943.1 DMQF01000030 HAO14052.1 QOIP01000004 RLU23301.1 DNUT01000056 HBI39512.1 DMQF01000478 HAO15711.1 GEBQ01006447 JAT33530.1 DNUT01000170 HBI39961.1 GBHO01002973 JAG40631.1 GEZM01041318 GEZM01041317 JAV80524.1 GANO01004275 JAB55596.1 GAMC01021037 GAMC01021036 JAB85519.1 LSMT01000554 PFX16333.1 DNUT01000297 HBI40414.1 GEZM01051381 GEZM01051380 JAV74995.1 DNUT01000374 HBI40695.1 GEHC01001026 JAV46619.1 GANO01001936 JAB57935.1 LSMT01001965 PFX11729.1 LSMT01000121 PFX26660.1 DNUT01000451 HBI40968.1 GAMC01021038 GAMC01021033 GAMC01021031 JAB85522.1 GEZM01074146 JAV64657.1 GEZM01061184 JAV70703.1 GEZM01061183 JAV70704.1 NWSH01001817 PCG70032.1 NWSH01000010 PCG80923.1 GANO01004361 JAB55510.1 GEZM01074147 JAV64656.1 LBMM01006440 KMQ90605.1 AAGJ04020328 AAZX01023386 AAZX01004038 GEZM01061182 JAV70705.1 GEZM01061185 JAV70702.1 GANO01004238 JAB55633.1 KZ150070 PZC74075.1 LSMT01000021 PFX32582.1 NWSH01006271 PCG63543.1 DMQF01000157 HAO14486.1 JXUM01065503 KQ562350 KXJ76100.1 GEZM01008907 JAV94684.1
Proteomes
Pfam
Interpro
IPR036691
Endo/exonu/phosph_ase_sf
+ More
IPR000477 RT_dom
IPR005135 Endo/exonuclease/phosphatase
IPR000639 Epox_hydrolase-like
IPR010497 Epoxide_hydro_N
IPR029058 AB_hydrolase
IPR001878 Znf_CCHC
IPR011011 Znf_FYVE_PHD
IPR021109 Peptidase_aspartic_dom_sf
IPR040676 DUF5641
IPR041373 RT_RNaseH
IPR036397 RNaseH_sf
IPR001584 Integrase_cat-core
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
IPR000477 RT_dom
IPR005135 Endo/exonuclease/phosphatase
IPR000639 Epox_hydrolase-like
IPR010497 Epoxide_hydro_N
IPR029058 AB_hydrolase
IPR001878 Znf_CCHC
IPR011011 Znf_FYVE_PHD
IPR021109 Peptidase_aspartic_dom_sf
IPR040676 DUF5641
IPR041373 RT_RNaseH
IPR036397 RNaseH_sf
IPR001584 Integrase_cat-core
IPR008042 Retrotrans_Pao
IPR012337 RNaseH-like_sf
SUPFAM
Gene 3D
ProteinModelPortal
A0A2A4JRM8
A0A2A4JPY6
A0A3S2P9A9
A0A3S2TB34
A0A1B6L6Y3
A0A0J7K589
+ More
A0A0J7N335 A0A0A9YIX2 A0A354GJG9 A0A1B6KD49 A0A3C1S1N3 A0A3C1S2P1 A0A1S3DM39 U5ENG8 A0A1W7R6E4 A0A1W7R6F6 A0A3L8D9C4 A0A3L8DQD7 A0A1W7R6K8 U5EQV9 A0A1W7R6J3 A0A3C1RY27 A0A3C1RYE8 A0A3L8DSB3 A0A354GES4 A0A3C1S360 A0A1B6MC85 A0A354GG23 A0A0A9Z9S6 A0A1Y1M3U5 U5EEL4 W8B6W6 A0A2B4RHP5 A0A354GHC6 A0A1Y1LSZ2 A0A354GI57 A0A1W7R628 U5EVC6 A0A2B4R4J4 A0A2B4SCV9 A0A354GIY0 A0A3B3BNA3 W8AIM4 A0A1Y1L0L4 A0A1Y1LAN8 A0A1Y1LAP1 A0A3P9M960 A0A2A4JF59 A0A3B3HIF3 A0A2A4KAV4 A0A3B3HCE9 U5EQB9 A0A3B3HNZ2 A0A1Y1KZ78 A0A0J7NDM1 A0A3P8PBK6 A0A3P9IXQ6 A0A3B3DM95 W4Z8W8 K7JAP1 A0A3B1JMQ8 K7JTK2 A0A3P9K1B3 A0A1Y1LFG1 A0A1Y1LAM2 U5ETR7 A0A2W1BK42 A0A2B4SVL4 A0A2A4IWQ4 A0A3C1RZS6 A0A182GI84 A0A1Y1N9S9
A0A0J7N335 A0A0A9YIX2 A0A354GJG9 A0A1B6KD49 A0A3C1S1N3 A0A3C1S2P1 A0A1S3DM39 U5ENG8 A0A1W7R6E4 A0A1W7R6F6 A0A3L8D9C4 A0A3L8DQD7 A0A1W7R6K8 U5EQV9 A0A1W7R6J3 A0A3C1RY27 A0A3C1RYE8 A0A3L8DSB3 A0A354GES4 A0A3C1S360 A0A1B6MC85 A0A354GG23 A0A0A9Z9S6 A0A1Y1M3U5 U5EEL4 W8B6W6 A0A2B4RHP5 A0A354GHC6 A0A1Y1LSZ2 A0A354GI57 A0A1W7R628 U5EVC6 A0A2B4R4J4 A0A2B4SCV9 A0A354GIY0 A0A3B3BNA3 W8AIM4 A0A1Y1L0L4 A0A1Y1LAN8 A0A1Y1LAP1 A0A3P9M960 A0A2A4JF59 A0A3B3HIF3 A0A2A4KAV4 A0A3B3HCE9 U5EQB9 A0A3B3HNZ2 A0A1Y1KZ78 A0A0J7NDM1 A0A3P8PBK6 A0A3P9IXQ6 A0A3B3DM95 W4Z8W8 K7JAP1 A0A3B1JMQ8 K7JTK2 A0A3P9K1B3 A0A1Y1LFG1 A0A1Y1LAM2 U5ETR7 A0A2W1BK42 A0A2B4SVL4 A0A2A4IWQ4 A0A3C1RZS6 A0A182GI84 A0A1Y1N9S9
Ontologies
Topology
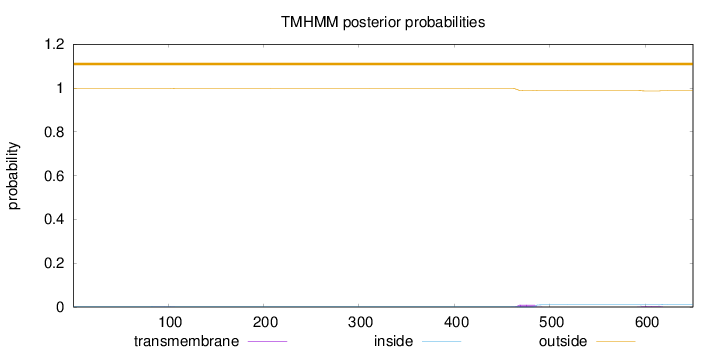
Length:
650
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.3132
Exp number, first 60 AAs:
0.000610000000000001
Total prob of N-in:
0.00235
outside
1 - 650
Population Genetic Test Statistics
Pi
239.864825
Theta
129.871034
Tajima's D
4.574526
CLR
0.285186
CSRT
0.9998000099995
Interpretation
Uncertain
