Gene
KWMTBOMO11959
Pre Gene Modal
BGIBMGA004381
Annotation
PREDICTED:_peroxisome_biogenesis_factor_10_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 3.613
Sequence
CDS
ATGGCTTTGCCAGTAGCACAACCTGCTGAAGTTCTACGTGCATGGCAAAAAGACGATCAATACGAAAAACAACTGGCCGATTCCATATCTAAACTGTTACCATTACAGCACGGTTCAAAAGCTATTCCAATATCCTCTTTGCTTTATAAATCATTTACAACCTTAAAAGACCTACAGACCCTAGGCGAAGAGTACTCTGGAATCGTCCAAGTAGATGATAGTTACCACAAACTGCCGAGTTACTACGCACGTTTAGCAAGCGTATTATTATCAACTTTTGGTGAAAATCTAACTCGAAGATTTGTAAATCATGTTGGAAAAAAAATTGAAACAAATCGATCACTGAGATCTGAAGCCCAAGATCTCTTTTTGATTGCAGTAAAGACTCTTGGTGATGTTTTGCCACAGATACAAAGTATTCACAGGGCATTATCTTACATCTGCGGAGGACCTTTACAGATAGGTAAAAATCTGACTGGCATTGATTATGTTCATGTTAGACCTGCTGCAACTGCTTCATATGCTCATTTAAATTTACTAGGCTACATAACATTACTGCATGCATTTGTATGCTGTGGACAAAGTGTTTACTTAGCGAAAAAACATTTGGACCAATTAAAAGAAATTCCCAATGAAGTGGATAACGAAAAGACATGTGTTGCATGCTTGGAGGAAATATTACAACCTTGTGTTCTTCAGTGTGGCCATATATTCTGCCATCAATGTTGCTATGGCGCACTAAACAGTTGTGCATTGTGCAGGACACCATTTACAAAGAATACTGTTGTGCCACTAATGAACTATTATCCTGGAGGTTAA
Protein
MALPVAQPAEVLRAWQKDDQYEKQLADSISKLLPLQHGSKAIPISSLLYKSFTTLKDLQTLGEEYSGIVQVDDSYHKLPSYYARLASVLLSTFGENLTRRFVNHVGKKIETNRSLRSEAQDLFLIAVKTLGDVLPQIQSIHRALSYICGGPLQIGKNLTGIDYVHVRPAATASYAHLNLLGYITLLHAFVCCGQSVYLAKKHLDQLKEIPNEVDNEKTCVACLEEILQPCVLQCGHIFCHQCCYGALNSCALCRTPFTKNTVVPLMNYYPGG
Summary
Uniprot
H9J4E2
A0A2A4JYQ9
A0A212FAN3
A0A2W1BAZ8
A0A0L7KNW6
A0A2H1W8H4
+ More
A0A0N1IGB0 A0A194Q577 A0A1B6DJM2 A0A336MIV3 A0A1J1IVX2 A0A1B0CSJ0 A0A1L8DSP0 A0A1B0D3U7 A0A0P4VQ13 A0A1B6MGH9 A0A069DRF6 A0A2R7WTF4 A0A224XPS7 B4PDA5 A0A0P4WIT5 A0A1I8MMI2 A0A0K8TT86 W8BX14 A0A1B0B8U6 A0A1B0A6W7 A0A1W4W623 A0A1A9URQ3 A0A1A9XB33 A0A0V0G6X4 A0A2M4AWP8 A0A182MIB7 A0A146LKP9 A0A0A9X2V9 A0A0K8THW6 B3NBC5 D3TPT9 U5EZB7 Q9W0D7 A0A0A1WM57 B3M8F1 A0A182TLP5 B4QMG3 A0A182PL40 W5JGW4 A0A182QUP1 A0A182UST9 A0A1I8PK36 B4HVV1 A0A232F8N7 A0A182WRB4 A0A182VT35 A0A182RKB5 K7IXV6 A0A182IDJ4 Q7QI38 A0A3B0KA22 A0A182T164 Q2LZX3 A0A088A4P5 A0A182NVE8 A0A182YQ34 B4LHM3 A0A310SIR6 B4IZJ9 A0A182LAB0 A0A2J7QVN4 A0A067R3D6 B4N2V2 A0A0L7R3C2 B4KWA1 A0A2A3ECF7 A0A154P3S6 A0A2P8Z684 A0A182J4R3 A0A139WMJ2 A0A2H8U1S7 E0VG81 A0A087SVF3 A0A0M8ZU16 A0A182H1E7 F5HIZ6 A0A1B6GG87 G3SYP6 A0A224YVM9 K1R1W6 A0A226E8G7 A0A2Y9E1T5 A0A131XZY1 J9M316 A0A2S2PZU2 A0A2S2PFY2 A0A182FK46 E2ATT3
A0A0N1IGB0 A0A194Q577 A0A1B6DJM2 A0A336MIV3 A0A1J1IVX2 A0A1B0CSJ0 A0A1L8DSP0 A0A1B0D3U7 A0A0P4VQ13 A0A1B6MGH9 A0A069DRF6 A0A2R7WTF4 A0A224XPS7 B4PDA5 A0A0P4WIT5 A0A1I8MMI2 A0A0K8TT86 W8BX14 A0A1B0B8U6 A0A1B0A6W7 A0A1W4W623 A0A1A9URQ3 A0A1A9XB33 A0A0V0G6X4 A0A2M4AWP8 A0A182MIB7 A0A146LKP9 A0A0A9X2V9 A0A0K8THW6 B3NBC5 D3TPT9 U5EZB7 Q9W0D7 A0A0A1WM57 B3M8F1 A0A182TLP5 B4QMG3 A0A182PL40 W5JGW4 A0A182QUP1 A0A182UST9 A0A1I8PK36 B4HVV1 A0A232F8N7 A0A182WRB4 A0A182VT35 A0A182RKB5 K7IXV6 A0A182IDJ4 Q7QI38 A0A3B0KA22 A0A182T164 Q2LZX3 A0A088A4P5 A0A182NVE8 A0A182YQ34 B4LHM3 A0A310SIR6 B4IZJ9 A0A182LAB0 A0A2J7QVN4 A0A067R3D6 B4N2V2 A0A0L7R3C2 B4KWA1 A0A2A3ECF7 A0A154P3S6 A0A2P8Z684 A0A182J4R3 A0A139WMJ2 A0A2H8U1S7 E0VG81 A0A087SVF3 A0A0M8ZU16 A0A182H1E7 F5HIZ6 A0A1B6GG87 G3SYP6 A0A224YVM9 K1R1W6 A0A226E8G7 A0A2Y9E1T5 A0A131XZY1 J9M316 A0A2S2PZU2 A0A2S2PFY2 A0A182FK46 E2ATT3
Pubmed
19121390
22118469
28756777
26227816
26354079
27129103
+ More
26334808 17994087 17550304 25315136 26369729 24495485 26823975 25401762 20353571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 22936249 20920257 23761445 28648823 20075255 12364791 14747013 17210077 15632085 25244985 20966253 24845553 29403074 18362917 19820115 20566863 26483478 28797301 22992520 20798317
26334808 17994087 17550304 25315136 26369729 24495485 26823975 25401762 20353571 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 22936249 20920257 23761445 28648823 20075255 12364791 14747013 17210077 15632085 25244985 20966253 24845553 29403074 18362917 19820115 20566863 26483478 28797301 22992520 20798317
EMBL
BABH01022022
NWSH01000415
PCG76550.1
AGBW02009448
OWR50788.1
KZ150208
+ More
PZC72228.1 JTDY01008009 KOB64800.1 ODYU01007025 SOQ49399.1 KQ460009 KPJ18470.1 KQ459460 KPJ00693.1 GEDC01011441 JAS25857.1 UFQT01000635 SSX25978.1 CVRI01000063 CRL04357.1 AJWK01026143 GFDF01004588 JAV09496.1 AJVK01023707 GDKW01001500 JAI55095.1 GEBQ01004972 JAT35005.1 GBGD01002409 JAC86480.1 KK855501 PTY22846.1 GFTR01004588 JAW11838.1 CM000159 EDW92853.1 GDRN01043000 JAI67005.1 GDAI01000478 JAI17125.1 GAMC01012811 JAB93744.1 JXJN01010060 GECL01002419 JAP03705.1 GGFK01011879 MBW45200.1 AXCM01003211 GDHC01011302 JAQ07327.1 GBHO01029276 GBHO01029275 GBHO01029274 JAG14328.1 JAG14329.1 JAG14330.1 GBRD01000638 JAG65183.1 CH954178 EDV50178.1 CCAG010023157 EZ423441 ADD19717.1 GANO01001082 JAB58789.1 AE014296 AY069290 AAF47512.1 AAL39435.1 AGB93960.1 GBXI01014370 JAC99921.1 CH902618 EDV38886.1 CM000363 CM002912 EDX08820.1 KMY96863.1 ADMH02001566 ETN62049.1 AXCN02002011 CH480817 EDW50066.1 NNAY01000686 OXU26972.1 APCN01004705 AAAB01008811 EAA04957.3 OUUW01000009 SPP84970.1 CH379069 EAL31164.3 CH940647 EDW69576.1 KQ760801 OAD58987.1 CH916366 EDV97774.1 NEVH01009776 PNF32643.1 KK852726 KDR17652.1 CH964062 EDW78691.1 KQ414663 KOC65276.1 CH933809 EDW18508.1 KZ288295 PBC28972.1 KQ434803 KZC05888.1 PYGN01000176 PSN52013.1 AXCP01007142 KQ971312 KYB29057.1 GFXV01007603 MBW19408.1 DS235132 EEB12387.1 KK112143 KFM56842.1 KQ435885 KOX69613.1 JXUM01022982 KQ560646 KXJ81457.1 EGK96257.1 GECZ01008446 JAS61323.1 GFPF01010502 MAA21648.1 JH816958 EKC27741.1 LNIX01000006 OXA53151.1 GEFM01004021 JAP71775.1 ABLF02026136 GGMS01001842 MBY71045.1 GGMR01015742 MBY28361.1 GL442691 EFN63166.1
PZC72228.1 JTDY01008009 KOB64800.1 ODYU01007025 SOQ49399.1 KQ460009 KPJ18470.1 KQ459460 KPJ00693.1 GEDC01011441 JAS25857.1 UFQT01000635 SSX25978.1 CVRI01000063 CRL04357.1 AJWK01026143 GFDF01004588 JAV09496.1 AJVK01023707 GDKW01001500 JAI55095.1 GEBQ01004972 JAT35005.1 GBGD01002409 JAC86480.1 KK855501 PTY22846.1 GFTR01004588 JAW11838.1 CM000159 EDW92853.1 GDRN01043000 JAI67005.1 GDAI01000478 JAI17125.1 GAMC01012811 JAB93744.1 JXJN01010060 GECL01002419 JAP03705.1 GGFK01011879 MBW45200.1 AXCM01003211 GDHC01011302 JAQ07327.1 GBHO01029276 GBHO01029275 GBHO01029274 JAG14328.1 JAG14329.1 JAG14330.1 GBRD01000638 JAG65183.1 CH954178 EDV50178.1 CCAG010023157 EZ423441 ADD19717.1 GANO01001082 JAB58789.1 AE014296 AY069290 AAF47512.1 AAL39435.1 AGB93960.1 GBXI01014370 JAC99921.1 CH902618 EDV38886.1 CM000363 CM002912 EDX08820.1 KMY96863.1 ADMH02001566 ETN62049.1 AXCN02002011 CH480817 EDW50066.1 NNAY01000686 OXU26972.1 APCN01004705 AAAB01008811 EAA04957.3 OUUW01000009 SPP84970.1 CH379069 EAL31164.3 CH940647 EDW69576.1 KQ760801 OAD58987.1 CH916366 EDV97774.1 NEVH01009776 PNF32643.1 KK852726 KDR17652.1 CH964062 EDW78691.1 KQ414663 KOC65276.1 CH933809 EDW18508.1 KZ288295 PBC28972.1 KQ434803 KZC05888.1 PYGN01000176 PSN52013.1 AXCP01007142 KQ971312 KYB29057.1 GFXV01007603 MBW19408.1 DS235132 EEB12387.1 KK112143 KFM56842.1 KQ435885 KOX69613.1 JXUM01022982 KQ560646 KXJ81457.1 EGK96257.1 GECZ01008446 JAS61323.1 GFPF01010502 MAA21648.1 JH816958 EKC27741.1 LNIX01000006 OXA53151.1 GEFM01004021 JAP71775.1 ABLF02026136 GGMS01001842 MBY71045.1 GGMR01015742 MBY28361.1 GL442691 EFN63166.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000037510
UP000053240
UP000053268
+ More
UP000183832 UP000092461 UP000092462 UP000002282 UP000095301 UP000092460 UP000092445 UP000192221 UP000078200 UP000092443 UP000075883 UP000008711 UP000092444 UP000000803 UP000007801 UP000075902 UP000000304 UP000075885 UP000000673 UP000075886 UP000075903 UP000095300 UP000001292 UP000215335 UP000076407 UP000075920 UP000075900 UP000002358 UP000075840 UP000007062 UP000268350 UP000075901 UP000001819 UP000005203 UP000075884 UP000076408 UP000008792 UP000001070 UP000075882 UP000235965 UP000027135 UP000007798 UP000053825 UP000009192 UP000242457 UP000076502 UP000245037 UP000075880 UP000007266 UP000009046 UP000054359 UP000053105 UP000069940 UP000249989 UP000007646 UP000005408 UP000198287 UP000248480 UP000007819 UP000069272 UP000000311
UP000183832 UP000092461 UP000092462 UP000002282 UP000095301 UP000092460 UP000092445 UP000192221 UP000078200 UP000092443 UP000075883 UP000008711 UP000092444 UP000000803 UP000007801 UP000075902 UP000000304 UP000075885 UP000000673 UP000075886 UP000075903 UP000095300 UP000001292 UP000215335 UP000076407 UP000075920 UP000075900 UP000002358 UP000075840 UP000007062 UP000268350 UP000075901 UP000001819 UP000005203 UP000075884 UP000076408 UP000008792 UP000001070 UP000075882 UP000235965 UP000027135 UP000007798 UP000053825 UP000009192 UP000242457 UP000076502 UP000245037 UP000075880 UP000007266 UP000009046 UP000054359 UP000053105 UP000069940 UP000249989 UP000007646 UP000005408 UP000198287 UP000248480 UP000007819 UP000069272 UP000000311
PRIDE
Interpro
SUPFAM
SSF46565
SSF46565
Gene 3D
CDD
ProteinModelPortal
H9J4E2
A0A2A4JYQ9
A0A212FAN3
A0A2W1BAZ8
A0A0L7KNW6
A0A2H1W8H4
+ More
A0A0N1IGB0 A0A194Q577 A0A1B6DJM2 A0A336MIV3 A0A1J1IVX2 A0A1B0CSJ0 A0A1L8DSP0 A0A1B0D3U7 A0A0P4VQ13 A0A1B6MGH9 A0A069DRF6 A0A2R7WTF4 A0A224XPS7 B4PDA5 A0A0P4WIT5 A0A1I8MMI2 A0A0K8TT86 W8BX14 A0A1B0B8U6 A0A1B0A6W7 A0A1W4W623 A0A1A9URQ3 A0A1A9XB33 A0A0V0G6X4 A0A2M4AWP8 A0A182MIB7 A0A146LKP9 A0A0A9X2V9 A0A0K8THW6 B3NBC5 D3TPT9 U5EZB7 Q9W0D7 A0A0A1WM57 B3M8F1 A0A182TLP5 B4QMG3 A0A182PL40 W5JGW4 A0A182QUP1 A0A182UST9 A0A1I8PK36 B4HVV1 A0A232F8N7 A0A182WRB4 A0A182VT35 A0A182RKB5 K7IXV6 A0A182IDJ4 Q7QI38 A0A3B0KA22 A0A182T164 Q2LZX3 A0A088A4P5 A0A182NVE8 A0A182YQ34 B4LHM3 A0A310SIR6 B4IZJ9 A0A182LAB0 A0A2J7QVN4 A0A067R3D6 B4N2V2 A0A0L7R3C2 B4KWA1 A0A2A3ECF7 A0A154P3S6 A0A2P8Z684 A0A182J4R3 A0A139WMJ2 A0A2H8U1S7 E0VG81 A0A087SVF3 A0A0M8ZU16 A0A182H1E7 F5HIZ6 A0A1B6GG87 G3SYP6 A0A224YVM9 K1R1W6 A0A226E8G7 A0A2Y9E1T5 A0A131XZY1 J9M316 A0A2S2PZU2 A0A2S2PFY2 A0A182FK46 E2ATT3
A0A0N1IGB0 A0A194Q577 A0A1B6DJM2 A0A336MIV3 A0A1J1IVX2 A0A1B0CSJ0 A0A1L8DSP0 A0A1B0D3U7 A0A0P4VQ13 A0A1B6MGH9 A0A069DRF6 A0A2R7WTF4 A0A224XPS7 B4PDA5 A0A0P4WIT5 A0A1I8MMI2 A0A0K8TT86 W8BX14 A0A1B0B8U6 A0A1B0A6W7 A0A1W4W623 A0A1A9URQ3 A0A1A9XB33 A0A0V0G6X4 A0A2M4AWP8 A0A182MIB7 A0A146LKP9 A0A0A9X2V9 A0A0K8THW6 B3NBC5 D3TPT9 U5EZB7 Q9W0D7 A0A0A1WM57 B3M8F1 A0A182TLP5 B4QMG3 A0A182PL40 W5JGW4 A0A182QUP1 A0A182UST9 A0A1I8PK36 B4HVV1 A0A232F8N7 A0A182WRB4 A0A182VT35 A0A182RKB5 K7IXV6 A0A182IDJ4 Q7QI38 A0A3B0KA22 A0A182T164 Q2LZX3 A0A088A4P5 A0A182NVE8 A0A182YQ34 B4LHM3 A0A310SIR6 B4IZJ9 A0A182LAB0 A0A2J7QVN4 A0A067R3D6 B4N2V2 A0A0L7R3C2 B4KWA1 A0A2A3ECF7 A0A154P3S6 A0A2P8Z684 A0A182J4R3 A0A139WMJ2 A0A2H8U1S7 E0VG81 A0A087SVF3 A0A0M8ZU16 A0A182H1E7 F5HIZ6 A0A1B6GG87 G3SYP6 A0A224YVM9 K1R1W6 A0A226E8G7 A0A2Y9E1T5 A0A131XZY1 J9M316 A0A2S2PZU2 A0A2S2PFY2 A0A182FK46 E2ATT3
PDB
5DIN
E-value=4.23734e-05,
Score=110
Ontologies
GO
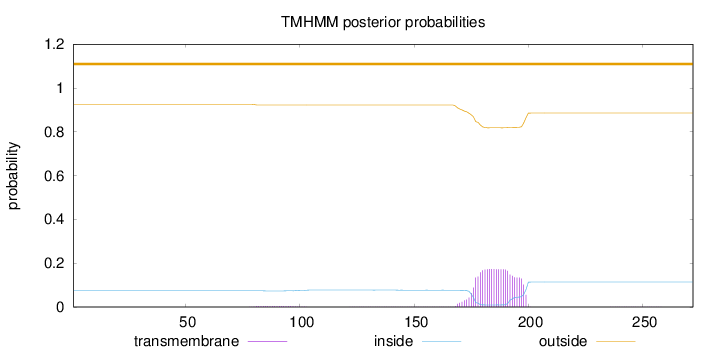
Topology
Length:
272
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
3.89309
Exp number, first 60 AAs:
0.00116
Total prob of N-in:
0.07374
outside
1 - 272
Population Genetic Test Statistics
Pi
271.695293
Theta
208.910771
Tajima's D
1.114881
CLR
0.043318
CSRT
0.690615469226539
Interpretation
Uncertain
