Gene
KWMTBOMO11951 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004476
Annotation
microvitellogenin-like_precursor_[Bombyx_mori]
Full name
Microvitellogenin
+ More
Low molecular mass 30 kDa lipoprotein 21G1
Low molecular mass 30 kDa lipoprotein 19G1
Low molecular 30 kDa lipoprotein PBMHPC-19
Low molecular 30 kDa lipoprotein PBMHPC-23
Low molecular 30 kDa lipoprotein PBMHP-6
Low molecular 30 kDa lipoprotein PBMHP-12
Low molecular 30 kDa lipoprotein PBMHPC-21
Low molecular mass 30 kDa lipoprotein 21G1
Low molecular mass 30 kDa lipoprotein 19G1
Low molecular 30 kDa lipoprotein PBMHPC-19
Low molecular 30 kDa lipoprotein PBMHPC-23
Low molecular 30 kDa lipoprotein PBMHP-6
Low molecular 30 kDa lipoprotein PBMHP-12
Low molecular 30 kDa lipoprotein PBMHPC-21
Location in the cell
Cytoplasmic Reliability : 1.072 Nuclear Reliability : 1.034
Sequence
CDS
ATGGTCCAAAACCTCTACGATGCCATTGTTTACGAAAACTACGACGAAGCGGTTTCCCAAGCCTTAAACTGTATGCGTTATGATCGACCCGATGTCATTAAATTAACTGTCGACAAGTTAATTCGGCAACAGAGAACATTTATTGCTGGTTTCGCATACAAACTGGGGGCTGCTGGGGGCGAAGATATTATAAGGTCCGAGTTCCCCCCGGCGTACAGAATGGTGCTAAACGATACTTTTATAAAATTCATCAACTACCAAGATCGTGTTTCCATAGAGTTAGGTATAACTCCAGACTTTGCCGATGACAAGAGGCTCTACGGCAACAACGATCATCCGCATAGCCAGAACTACCATTGGAAGCTCGAGCCCGTGATGGATGGTAAAAAGATTTATTTCAAGATACGTCATCGCGTAGAAGAAAGGTACTTGAAGCTGGCCAACAACGCGGACCGCGACGGCGACTATCAAGCGTATGCTTCCACAAACTCAGACACGAACAGGCACCTGTGGTCGTTCCAGCCTGTGATGTACAACGGGGAGCTCTTGTTCTTCGTTATCAATCGTGCGCATTTCCAAGCGCTGAAGTTCGGCCGACGCGTATATGATGGTAGTCGCAACCTTTACAGTCAAGGTGGAAACTTTGCCGCCCACCCTGAACTTAACGGCTGGACCATCGAAGCTTTCGGAGACAGTTCCAAATTTGTAATTATCAGGGACTAG
Protein
MVQNLYDAIVYENYDEAVSQALNCMRYDRPDVIKLTVDKLIRQQRTFIAGFAYKLGAAGGEDIIRSEFPPAYRMVLNDTFIKFINYQDRVSIELGITPDFADDKRLYGNNDHPHSQNYHWKLEPVMDGKKIYFKIRHRVEERYLKLANNADRDGDYQAYASTNSDTNRHLWSFQPVMYNGELLFFVINRAHFQALKFGRRVYDGSRNLYSQGGNFAAHPELNGWTIEAFGDSSKFVIIRD
Summary
Miscellaneous
This lipoprotein belongs to the group of structurally related '30 kDa proteins' that comprise major protein components of the fifth (and last) instar larvae and of pupae.
Biosynthesis occurs in a stage-dependent fashion in the fat body.
Biosynthesis occurs in a stage-dependent fashion in the fat body.
Similarity
Belongs to the 30 kDa lipoprotein family.
Keywords
Signal
Complete proteome
Lipoprotein
Reference proteome
3D-structure
Secreted
Feature
chain Microvitellogenin
Uniprot
H9B459
H9J4N6
H9JXA6
H9B462
H9JVX1
P19616
+ More
Q0VJU3 H9J4M4 Q05432 H9J4M3 H9J4M7 H9J4M5 H9J4M8 A0A0B5Z504 H9B453 H9J4N1 H9J4N4 H9J4E8 H9JX32 H9J4E6 Q75RW3 H9J4N3 H9JX33 Q6Q0S8 H9J4F5 D4QGB9 H9J4F6 Q00801 Q00802 C7A8A3 H9J4L1 H9J4G3 H9J4L6 A7LIK7 P09336 D4QGC0 H9J4E7 H9J4N5 H9B458 E5EVW2 P09338 H9B443 P09334 C7A8A2 Q17185 H9J4G4 H9B444 H9B436 H9B450 H9J4G0 P09335 H9J4L5 H9B437 H9J4F9 H9B440 H9J4G1 H9J4G5 A0A2W1BZD4 A0A3G1LBE0 H9B449 H9JMX4 H9B451 A0A385HW64 Q76IB6 H9B447 E5EVW3 H9B448 A0A194QYV5 H9B445 H9B455 A0A2A4K904 H9JJC0 H9JJ72 A0A2W1BV38 H9B460 A0A3G1LBD4 B5BSX5 H9B454 H9JF68 Q2PQU4 A0A194Q636 H9JL03 H9J4L7 A0A2W1BPF5 P09337 A0A2A4JTT3 H9J4G2 A0A2H1WST8 A0A2A4IYY5 H9B457
Q0VJU3 H9J4M4 Q05432 H9J4M3 H9J4M7 H9J4M5 H9J4M8 A0A0B5Z504 H9B453 H9J4N1 H9J4N4 H9J4E8 H9JX32 H9J4E6 Q75RW3 H9J4N3 H9JX33 Q6Q0S8 H9J4F5 D4QGB9 H9J4F6 Q00801 Q00802 C7A8A3 H9J4L1 H9J4G3 H9J4L6 A7LIK7 P09336 D4QGC0 H9J4E7 H9J4N5 H9B458 E5EVW2 P09338 H9B443 P09334 C7A8A2 Q17185 H9J4G4 H9B444 H9B436 H9B450 H9J4G0 P09335 H9J4L5 H9B437 H9J4F9 H9B440 H9J4G1 H9J4G5 A0A2W1BZD4 A0A3G1LBE0 H9B449 H9JMX4 H9B451 A0A385HW64 Q76IB6 H9B447 E5EVW3 H9B448 A0A194QYV5 H9B445 H9B455 A0A2A4K904 H9JJC0 H9JJ72 A0A2W1BV38 H9B460 A0A3G1LBD4 B5BSX5 H9B454 H9JF68 Q2PQU4 A0A194Q636 H9JL03 H9J4L7 A0A2W1BPF5 P09337 A0A2A4JTT3 H9J4G2 A0A2H1WST8 A0A2A4IYY5 H9B457
Pubmed
EMBL
JN977544
AFC87824.1
BABH01022014
BABH01035696
JN977547
AFC87827.1
+ More
BABH01040631 M28820 J03768 AM293331 CAL25136.1 BABH01021994 JN977526 AFC87806.1 D12523 BAA02093.1 BABH01021988 BABH01021999 BABH01022000 BABH01021998 JN977527 AFC87807.1 KM926620 AJI02615.1 JN977538 AFC87818.1 BABH01022003 BABH01022005 BABH01022004 BABH01043627 BABH01022007 AB158646 BAD07027.1 BABH01043629 AY568957 AAS75435.1 BABH01021941 JN977524 AFC87804.1 AB553283 BAJ04787.1 BABH01021937 JN977519 AFC87799.1 X54734 X54736 FJ214661 ACO35751.1 BABH01021908 BABH01021929 BABH01021930 EU024295 ABS71120.1 X07554 AB553284 BAJ04788.1 BABH01022006 BABH01022013 JN977543 AFC87823.1 BABH01021936 HM012806 JN977520 ADQ89805.1 AFC87800.1 X07556 JN977528 AFC87808.1 X07552 FJ214660 ACO35750.1 X54735 CAA38532.1 BABH01021925 JN977523 AFC87803.1 JN977529 AFC87809.1 JN977521 AFC87801.1 JN977535 AFC87815.1 BABH01021933 BABH01021934 X07553 JN977522 AFC87802.1 BABH01021935 JN977525 AFC87805.1 BABH01021931 BABH01021932 BABH01021921 KZ149893 PZC78944.1 MF497270 AUB45122.1 JN977534 AFC87814.1 BABH01028980 JN977536 AFC87816.1 MG799571 AXY55009.1 AB098260 BAC82630.1 BABH01026781 JN977532 AFC87812.1 HM012807 ADQ89806.1 JN977533 AFC87813.1 KQ460930 KPJ10718.1 BABH01028979 JN977530 AFC87810.1 JN977540 AFC87820.1 NWSH01000017 PCG80765.1 BABH01036437 JN977541 AFC87821.1 PZC78918.1 JN977545 AFC87825.1 MF497269 AUB45121.1 AB436163 BAG70410.1 JN977539 AFC87819.1 BABH01026782 JN977537 AFC87817.1 BABH01011413 DQ306881 JN977531 ABC18268.1 AFC87811.1 KQ459582 KPI98870.1 KZ149940 PZC76929.1 X07555 NWSH01000685 PCG74802.1 ODYU01010790 SOQ56108.1 NWSH01005147 PCG64400.1 JN977542 AFC87822.1
BABH01040631 M28820 J03768 AM293331 CAL25136.1 BABH01021994 JN977526 AFC87806.1 D12523 BAA02093.1 BABH01021988 BABH01021999 BABH01022000 BABH01021998 JN977527 AFC87807.1 KM926620 AJI02615.1 JN977538 AFC87818.1 BABH01022003 BABH01022005 BABH01022004 BABH01043627 BABH01022007 AB158646 BAD07027.1 BABH01043629 AY568957 AAS75435.1 BABH01021941 JN977524 AFC87804.1 AB553283 BAJ04787.1 BABH01021937 JN977519 AFC87799.1 X54734 X54736 FJ214661 ACO35751.1 BABH01021908 BABH01021929 BABH01021930 EU024295 ABS71120.1 X07554 AB553284 BAJ04788.1 BABH01022006 BABH01022013 JN977543 AFC87823.1 BABH01021936 HM012806 JN977520 ADQ89805.1 AFC87800.1 X07556 JN977528 AFC87808.1 X07552 FJ214660 ACO35750.1 X54735 CAA38532.1 BABH01021925 JN977523 AFC87803.1 JN977529 AFC87809.1 JN977521 AFC87801.1 JN977535 AFC87815.1 BABH01021933 BABH01021934 X07553 JN977522 AFC87802.1 BABH01021935 JN977525 AFC87805.1 BABH01021931 BABH01021932 BABH01021921 KZ149893 PZC78944.1 MF497270 AUB45122.1 JN977534 AFC87814.1 BABH01028980 JN977536 AFC87816.1 MG799571 AXY55009.1 AB098260 BAC82630.1 BABH01026781 JN977532 AFC87812.1 HM012807 ADQ89806.1 JN977533 AFC87813.1 KQ460930 KPJ10718.1 BABH01028979 JN977530 AFC87810.1 JN977540 AFC87820.1 NWSH01000017 PCG80765.1 BABH01036437 JN977541 AFC87821.1 PZC78918.1 JN977545 AFC87825.1 MF497269 AUB45121.1 AB436163 BAG70410.1 JN977539 AFC87819.1 BABH01026782 JN977537 AFC87817.1 BABH01011413 DQ306881 JN977531 ABC18268.1 AFC87811.1 KQ459582 KPI98870.1 KZ149940 PZC76929.1 X07555 NWSH01000685 PCG74802.1 ODYU01010790 SOQ56108.1 NWSH01005147 PCG64400.1 JN977542 AFC87822.1
Proteomes
PRIDE
H9B459
H9J4N6
H9JXA6
H9B462
H9J4M4
Q05432
+ More
H9J4M3 H9J4M7 H9J4M5 H9J4M8 H9B453 H9J4N1 H9J4N4 H9J4E8 H9JX32 H9J4E6 Q75RW3 H9J4N3 H9JX33 Q6Q0S8 H9J4F5 D4QGB9 H9J4F6 Q00801 Q00802 C7A8A3 H9J4L1 H9J4G3 H9J4L6 A7LIK7 P09336 D4QGC0 H9J4E7 H9J4N5 H9B458 E5EVW2 P09338 P09334 C7A8A2 Q17185 H9J4G4 H9B444 H9B436 H9B450 H9J4G0 P09335 H9J4L5 H9B437 H9J4F9 H9B440 H9J4G1 H9J4G5 H9B449 H9JMX4 H9B451 H9B447 E5EVW3 H9B448 H9B445 H9B455 H9JJC0 H9JJ72 H9B460 B5BSX5 H9B454 H9JF68 Q2PQU4 H9JL03 H9J4L7 P09337 H9J4G2 H9B457
H9J4M3 H9J4M7 H9J4M5 H9J4M8 H9B453 H9J4N1 H9J4N4 H9J4E8 H9JX32 H9J4E6 Q75RW3 H9J4N3 H9JX33 Q6Q0S8 H9J4F5 D4QGB9 H9J4F6 Q00801 Q00802 C7A8A3 H9J4L1 H9J4G3 H9J4L6 A7LIK7 P09336 D4QGC0 H9J4E7 H9J4N5 H9B458 E5EVW2 P09338 P09334 C7A8A2 Q17185 H9J4G4 H9B444 H9B436 H9B450 H9J4G0 P09335 H9J4L5 H9B437 H9J4F9 H9B440 H9J4G1 H9J4G5 H9B449 H9JMX4 H9B451 H9B447 E5EVW3 H9B448 H9B445 H9B455 H9JJC0 H9JJ72 H9B460 B5BSX5 H9B454 H9JF68 Q2PQU4 H9JL03 H9J4L7 P09337 H9J4G2 H9B457
Pfam
PF03260 Lipoprotein_11
SUPFAM
SSF50370
SSF50370
Gene 3D
ProteinModelPortal
H9B459
H9J4N6
H9JXA6
H9B462
H9JVX1
P19616
+ More
Q0VJU3 H9J4M4 Q05432 H9J4M3 H9J4M7 H9J4M5 H9J4M8 A0A0B5Z504 H9B453 H9J4N1 H9J4N4 H9J4E8 H9JX32 H9J4E6 Q75RW3 H9J4N3 H9JX33 Q6Q0S8 H9J4F5 D4QGB9 H9J4F6 Q00801 Q00802 C7A8A3 H9J4L1 H9J4G3 H9J4L6 A7LIK7 P09336 D4QGC0 H9J4E7 H9J4N5 H9B458 E5EVW2 P09338 H9B443 P09334 C7A8A2 Q17185 H9J4G4 H9B444 H9B436 H9B450 H9J4G0 P09335 H9J4L5 H9B437 H9J4F9 H9B440 H9J4G1 H9J4G5 A0A2W1BZD4 A0A3G1LBE0 H9B449 H9JMX4 H9B451 A0A385HW64 Q76IB6 H9B447 E5EVW3 H9B448 A0A194QYV5 H9B445 H9B455 A0A2A4K904 H9JJC0 H9JJ72 A0A2W1BV38 H9B460 A0A3G1LBD4 B5BSX5 H9B454 H9JF68 Q2PQU4 A0A194Q636 H9JL03 H9J4L7 A0A2W1BPF5 P09337 A0A2A4JTT3 H9J4G2 A0A2H1WST8 A0A2A4IYY5 H9B457
Q0VJU3 H9J4M4 Q05432 H9J4M3 H9J4M7 H9J4M5 H9J4M8 A0A0B5Z504 H9B453 H9J4N1 H9J4N4 H9J4E8 H9JX32 H9J4E6 Q75RW3 H9J4N3 H9JX33 Q6Q0S8 H9J4F5 D4QGB9 H9J4F6 Q00801 Q00802 C7A8A3 H9J4L1 H9J4G3 H9J4L6 A7LIK7 P09336 D4QGC0 H9J4E7 H9J4N5 H9B458 E5EVW2 P09338 H9B443 P09334 C7A8A2 Q17185 H9J4G4 H9B444 H9B436 H9B450 H9J4G0 P09335 H9J4L5 H9B437 H9J4F9 H9B440 H9J4G1 H9J4G5 A0A2W1BZD4 A0A3G1LBE0 H9B449 H9JMX4 H9B451 A0A385HW64 Q76IB6 H9B447 E5EVW3 H9B448 A0A194QYV5 H9B445 H9B455 A0A2A4K904 H9JJC0 H9JJ72 A0A2W1BV38 H9B460 A0A3G1LBD4 B5BSX5 H9B454 H9JF68 Q2PQU4 A0A194Q636 H9JL03 H9J4L7 A0A2W1BPF5 P09337 A0A2A4JTT3 H9J4G2 A0A2H1WST8 A0A2A4IYY5 H9B457
PDB
4IY9
E-value=1.22061e-27,
Score=304
Ontologies
GO
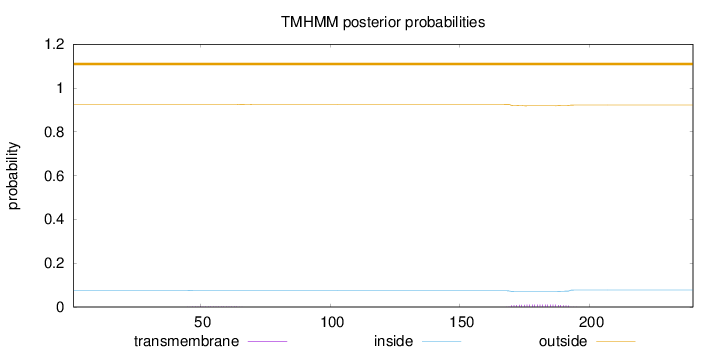
Topology
Subcellular location
Length:
240
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.27036
Exp number, first 60 AAs:
0.03269
Total prob of N-in:
0.07651
outside
1 - 240
Population Genetic Test Statistics
Pi
434.435205
Theta
212.81842
Tajima's D
3.278574
CLR
0.277705
CSRT
0.989200539973001
Interpretation
Uncertain
