Gene
KWMTBOMO11939
Pre Gene Modal
BGIBMGA004472
Annotation
PREDICTED:_uncharacterized_protein_LOC105380908_[Plutella_xylostella]
Location in the cell
Nuclear Reliability : 3.597
Sequence
CDS
ATGTCATACGAATCGGACATTAATAAAAGTGAAGTGGAGCAGATTACGGTTACATCGAGAATAGCCGAGTTCTGGGAGGATCAACCCAGGACGTGGTTCATACATGCAGAGTCAGTTCTTTTTAATCAAAAATTATCGGACGACGCCAAGTACCATCTCGTCATTGCCAAACTGGGTAAAAACGTGGTCAGTCAAGTGACAGATATATTAATAAAGCCGCCGAAAGAAAAAAAATATGACACACTCAAGGCCAGACTTTTAGCAATATACGAGGAATCGGAAAGCCACCAGTTGCAGAAGCTCATAGGGGAAATGGAACTGGGCGATCAGAAACCTTCGCAGCTGCTTCGTCGAATGAAAGATTTAGCCCGCGATAAAATCAGCGACGAAACCTTACTCCTTTTGTGGCAGAACCACCTACCAACCTCCGTCCGTGCAGTATTAGTAGTGACGGATAGCAAGGATATCAATACCCTGGCTTCCGTAGCCGACAAGGTAATGGAGACAACCAGGCCAATCTCCATAGGCGAGGTCAATACGGCAGATACAGCTACCTTAATATCGCAAATAGCCAGAATTAATATTCGCCTCGATGAGATCCAGTCGCAGAGGAATGGCCAACGCAGAGAACGGTACAGATACGGATTCCGTGGGCGCTATGCAAACAGAGCGCGATCGAGTTCCCGTGGTCGCCGTAATACGACACCCCATCGCTCAGCAGCTCGTGACGATTGGATGTGCTTCTATCACTACAGATTCAAGGAGCAGGCGCATAAATGCGTGCCGCCGTGTGCGTGGGAGAAGAAGCAGAACCAGTCGGGAAACTAA
Protein
MSYESDINKSEVEQITVTSRIAEFWEDQPRTWFIHAESVLFNQKLSDDAKYHLVIAKLGKNVVSQVTDILIKPPKEKKYDTLKARLLAIYEESESHQLQKLIGEMELGDQKPSQLLRRMKDLARDKISDETLLLLWQNHLPTSVRAVLVVTDSKDINTLASVADKVMETTRPISIGEVNTADTATLISQIARINIRLDEIQSQRNGQRRERYRYGFRGRYANRARSSSRGRRNTTPHRSAARDDWMCFYHYRFKEQAHKCVPPCAWEKKQNQSGN
Summary
Uniprot
H9J4N2
A0A2W1BAJ4
A0A2A4JUK3
A0A2H1WB17
A0A2H1V5N9
A0A1E1WLV2
+ More
A0A0L7LFW5 A0A2A4JI65 A0A0L7LSS0 A0A0L7L5A2 T1I831 T1HGU2 A0A0L7LFF1 A0A151JF00 A0A2W1BA89 T1IE85 A0A0J7K6L8 A0A0A9YNN7 A0A151K1Q2 A0A1Y1MA44 D7GYF3 A0A139WA14 D7EJX5 A0A0J7K6V1 A0A0K3CPY2 D6WTN9 T1HU96 A0A139W8G7 A0A151K2U9 A0A1I8Q981 A0A2A4J536 K7K066 A0A2L2YSA7 A0A0A9Z2L8 T1IE13 A0A139WPP8 A0A1I8NYN8 A0A139WPG8 A0A026VX87 A0A2A4K2H4 A0A026VRX8 A0A2G8JL30 A0A151JR52 A0A182G9J3 A0A131YM78 A0A151IIJ9 A0A0L7QK36 A0A182H3L9 A0A131Z7F2 A0A151K0A8 A0A151K1K5 A0A224XWW2 A0A1Y1MVP9 A0A0J7K6V3 A0A0K8SFH9 A0A2J7RGP3 K7JX19 A0A2L2YTR5 A0A151IZ47 A0A2J7QH56 A0A1I8Q298 A0A154PMS5 A0A2J7PML8 A0A154NZG0 A0A0N0BJL3 A0A0J7K6I6 A0A0L7QJB5 A0A085NTM8 A0A151ITG2 A0A0P8Y533 A0A085NBV0 A0A182H518 A0A0J7K7T5 A0A2I4C6S2 A0A085N631 A0A0J7NAK0 A0A085M716 A0A182GMM9 A0A147BQI1 W4YWZ1 A0A2J7QJD3 A0A0J7KD10 A0A2G8L3A3 A0A0P8ZL89 A0A0K8SB41
A0A0L7LFW5 A0A2A4JI65 A0A0L7LSS0 A0A0L7L5A2 T1I831 T1HGU2 A0A0L7LFF1 A0A151JF00 A0A2W1BA89 T1IE85 A0A0J7K6L8 A0A0A9YNN7 A0A151K1Q2 A0A1Y1MA44 D7GYF3 A0A139WA14 D7EJX5 A0A0J7K6V1 A0A0K3CPY2 D6WTN9 T1HU96 A0A139W8G7 A0A151K2U9 A0A1I8Q981 A0A2A4J536 K7K066 A0A2L2YSA7 A0A0A9Z2L8 T1IE13 A0A139WPP8 A0A1I8NYN8 A0A139WPG8 A0A026VX87 A0A2A4K2H4 A0A026VRX8 A0A2G8JL30 A0A151JR52 A0A182G9J3 A0A131YM78 A0A151IIJ9 A0A0L7QK36 A0A182H3L9 A0A131Z7F2 A0A151K0A8 A0A151K1K5 A0A224XWW2 A0A1Y1MVP9 A0A0J7K6V3 A0A0K8SFH9 A0A2J7RGP3 K7JX19 A0A2L2YTR5 A0A151IZ47 A0A2J7QH56 A0A1I8Q298 A0A154PMS5 A0A2J7PML8 A0A154NZG0 A0A0N0BJL3 A0A0J7K6I6 A0A0L7QJB5 A0A085NTM8 A0A151ITG2 A0A0P8Y533 A0A085NBV0 A0A182H518 A0A0J7K7T5 A0A2I4C6S2 A0A085N631 A0A0J7NAK0 A0A085M716 A0A182GMM9 A0A147BQI1 W4YWZ1 A0A2J7QJD3 A0A0J7KD10 A0A2G8L3A3 A0A0P8ZL89 A0A0K8SB41
Pubmed
EMBL
BABH01022003
KZ150191
PZC72412.1
NWSH01000589
PCG75479.1
ODYU01007352
+ More
SOQ50042.1 ODYU01000815 SOQ36165.1 GDQN01003070 JAT87984.1 JTDY01001300 KOB74269.1 NWSH01001424 PCG71274.1 JTDY01000162 KOB78510.1 JTDY01002934 KOB70464.1 ACPB03012735 ACPB03011651 KOB74268.1 KQ979024 KYN24351.1 KZ150227 PZC72038.1 ACPB03034542 LBMM01012633 KMQ86093.1 GBHO01008962 JAG34642.1 LKEX01009256 KYN50060.1 GEZM01036621 JAV82564.1 GG695519 EFA13383.1 KQ971745 KYB24749.1 KQ971759 EFA12902.2 LBMM01012923 LBMM01012917 KMQ85926.1 KMQ85930.1 LN877232 CTR11709.1 KQ971355 EFA07205.2 ACPB03016217 KQ973343 KXZ75568.1 LKEY01014428 KYN50445.1 NWSH01003151 PCG66856.1 AAZX01005161 IAAA01053498 IAAA01053499 LAA11042.1 GBHO01005513 JAG38091.1 ACPB03037918 KQ971307 KYB29887.1 KYB29888.1 KK107653 EZA48265.1 NWSH01000267 PCG77880.1 KK111495 EZA46390.1 MRZV01001674 PIK36447.1 KQ978645 KYN29457.1 JXUM01049417 KQ561603 KXJ78027.1 GEDV01008183 JAP80374.1 KQ977516 KYN02153.1 KQ414987 KOC58988.1 JXUM01107723 KQ565168 KXJ71261.1 GEDV01001795 JAP86762.1 KQ981301 KYN43041.1 LKEX01008659 KYN50015.1 GFTR01003955 JAW12471.1 GEZM01019580 GEZM01019578 JAV89619.1 LBMM01012479 KMQ86203.1 GBRD01013927 JAG51899.1 NEVH01003759 PNF40011.1 IAAA01048245 LAA10725.1 KQ980726 KYN14011.1 NEVH01014357 PNF27922.1 KQ434992 KZC13185.1 NEVH01023981 PNF17578.1 KQ434784 KZC04993.1 KQ435713 KOX79334.1 LBMM01013103 KMQ85816.1 LHQN01021927 LHQN01025379 KOC58628.1 KOC58659.1 KL367475 KFD72824.1 KQ981018 KYN10257.1 CH905996 KPU81830.1 KL367519 KFD66946.1 JXUM01111110 KQ565471 KXJ70931.1 LBMM01012075 KMQ86422.1 KL367547 KFD64927.1 LBMM01007548 KMQ89630.1 KL363221 KFD53012.1 JXUM01074757 KQ562849 KXJ75012.1 GEGO01002739 JAR92665.1 AAGJ04150373 NEVH01013558 PNF28672.1 LBMM01009142 KMQ88358.1 MRZV01001127 MRZV01000236 PIK40395.1 PIK54731.1 CH902629 KPU75574.1 GBRD01015841 JAG49985.1
SOQ50042.1 ODYU01000815 SOQ36165.1 GDQN01003070 JAT87984.1 JTDY01001300 KOB74269.1 NWSH01001424 PCG71274.1 JTDY01000162 KOB78510.1 JTDY01002934 KOB70464.1 ACPB03012735 ACPB03011651 KOB74268.1 KQ979024 KYN24351.1 KZ150227 PZC72038.1 ACPB03034542 LBMM01012633 KMQ86093.1 GBHO01008962 JAG34642.1 LKEX01009256 KYN50060.1 GEZM01036621 JAV82564.1 GG695519 EFA13383.1 KQ971745 KYB24749.1 KQ971759 EFA12902.2 LBMM01012923 LBMM01012917 KMQ85926.1 KMQ85930.1 LN877232 CTR11709.1 KQ971355 EFA07205.2 ACPB03016217 KQ973343 KXZ75568.1 LKEY01014428 KYN50445.1 NWSH01003151 PCG66856.1 AAZX01005161 IAAA01053498 IAAA01053499 LAA11042.1 GBHO01005513 JAG38091.1 ACPB03037918 KQ971307 KYB29887.1 KYB29888.1 KK107653 EZA48265.1 NWSH01000267 PCG77880.1 KK111495 EZA46390.1 MRZV01001674 PIK36447.1 KQ978645 KYN29457.1 JXUM01049417 KQ561603 KXJ78027.1 GEDV01008183 JAP80374.1 KQ977516 KYN02153.1 KQ414987 KOC58988.1 JXUM01107723 KQ565168 KXJ71261.1 GEDV01001795 JAP86762.1 KQ981301 KYN43041.1 LKEX01008659 KYN50015.1 GFTR01003955 JAW12471.1 GEZM01019580 GEZM01019578 JAV89619.1 LBMM01012479 KMQ86203.1 GBRD01013927 JAG51899.1 NEVH01003759 PNF40011.1 IAAA01048245 LAA10725.1 KQ980726 KYN14011.1 NEVH01014357 PNF27922.1 KQ434992 KZC13185.1 NEVH01023981 PNF17578.1 KQ434784 KZC04993.1 KQ435713 KOX79334.1 LBMM01013103 KMQ85816.1 LHQN01021927 LHQN01025379 KOC58628.1 KOC58659.1 KL367475 KFD72824.1 KQ981018 KYN10257.1 CH905996 KPU81830.1 KL367519 KFD66946.1 JXUM01111110 KQ565471 KXJ70931.1 LBMM01012075 KMQ86422.1 KL367547 KFD64927.1 LBMM01007548 KMQ89630.1 KL363221 KFD53012.1 JXUM01074757 KQ562849 KXJ75012.1 GEGO01002739 JAR92665.1 AAGJ04150373 NEVH01013558 PNF28672.1 LBMM01009142 KMQ88358.1 MRZV01001127 MRZV01000236 PIK40395.1 PIK54731.1 CH902629 KPU75574.1 GBRD01015841 JAG49985.1
Proteomes
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9J4N2
A0A2W1BAJ4
A0A2A4JUK3
A0A2H1WB17
A0A2H1V5N9
A0A1E1WLV2
+ More
A0A0L7LFW5 A0A2A4JI65 A0A0L7LSS0 A0A0L7L5A2 T1I831 T1HGU2 A0A0L7LFF1 A0A151JF00 A0A2W1BA89 T1IE85 A0A0J7K6L8 A0A0A9YNN7 A0A151K1Q2 A0A1Y1MA44 D7GYF3 A0A139WA14 D7EJX5 A0A0J7K6V1 A0A0K3CPY2 D6WTN9 T1HU96 A0A139W8G7 A0A151K2U9 A0A1I8Q981 A0A2A4J536 K7K066 A0A2L2YSA7 A0A0A9Z2L8 T1IE13 A0A139WPP8 A0A1I8NYN8 A0A139WPG8 A0A026VX87 A0A2A4K2H4 A0A026VRX8 A0A2G8JL30 A0A151JR52 A0A182G9J3 A0A131YM78 A0A151IIJ9 A0A0L7QK36 A0A182H3L9 A0A131Z7F2 A0A151K0A8 A0A151K1K5 A0A224XWW2 A0A1Y1MVP9 A0A0J7K6V3 A0A0K8SFH9 A0A2J7RGP3 K7JX19 A0A2L2YTR5 A0A151IZ47 A0A2J7QH56 A0A1I8Q298 A0A154PMS5 A0A2J7PML8 A0A154NZG0 A0A0N0BJL3 A0A0J7K6I6 A0A0L7QJB5 A0A085NTM8 A0A151ITG2 A0A0P8Y533 A0A085NBV0 A0A182H518 A0A0J7K7T5 A0A2I4C6S2 A0A085N631 A0A0J7NAK0 A0A085M716 A0A182GMM9 A0A147BQI1 W4YWZ1 A0A2J7QJD3 A0A0J7KD10 A0A2G8L3A3 A0A0P8ZL89 A0A0K8SB41
A0A0L7LFW5 A0A2A4JI65 A0A0L7LSS0 A0A0L7L5A2 T1I831 T1HGU2 A0A0L7LFF1 A0A151JF00 A0A2W1BA89 T1IE85 A0A0J7K6L8 A0A0A9YNN7 A0A151K1Q2 A0A1Y1MA44 D7GYF3 A0A139WA14 D7EJX5 A0A0J7K6V1 A0A0K3CPY2 D6WTN9 T1HU96 A0A139W8G7 A0A151K2U9 A0A1I8Q981 A0A2A4J536 K7K066 A0A2L2YSA7 A0A0A9Z2L8 T1IE13 A0A139WPP8 A0A1I8NYN8 A0A139WPG8 A0A026VX87 A0A2A4K2H4 A0A026VRX8 A0A2G8JL30 A0A151JR52 A0A182G9J3 A0A131YM78 A0A151IIJ9 A0A0L7QK36 A0A182H3L9 A0A131Z7F2 A0A151K0A8 A0A151K1K5 A0A224XWW2 A0A1Y1MVP9 A0A0J7K6V3 A0A0K8SFH9 A0A2J7RGP3 K7JX19 A0A2L2YTR5 A0A151IZ47 A0A2J7QH56 A0A1I8Q298 A0A154PMS5 A0A2J7PML8 A0A154NZG0 A0A0N0BJL3 A0A0J7K6I6 A0A0L7QJB5 A0A085NTM8 A0A151ITG2 A0A0P8Y533 A0A085NBV0 A0A182H518 A0A0J7K7T5 A0A2I4C6S2 A0A085N631 A0A0J7NAK0 A0A085M716 A0A182GMM9 A0A147BQI1 W4YWZ1 A0A2J7QJD3 A0A0J7KD10 A0A2G8L3A3 A0A0P8ZL89 A0A0K8SB41
Ontologies
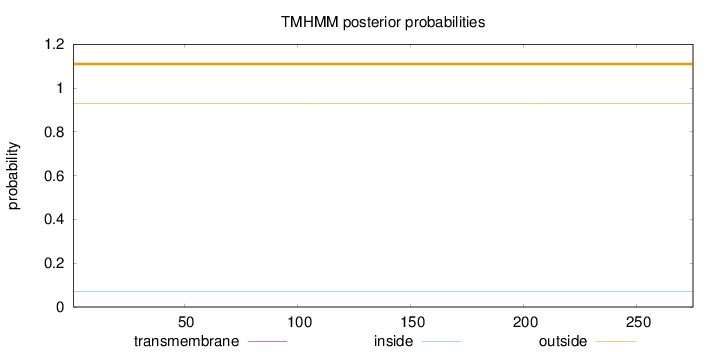
Topology
Length:
275
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00155
Exp number, first 60 AAs:
0.00027
Total prob of N-in:
0.07111
outside
1 - 275
Population Genetic Test Statistics
Pi
44.191235
Theta
87.623879
Tajima's D
-1.456484
CLR
0
CSRT
0.0670466476676166
Interpretation
Uncertain
