Gene
KWMTBOMO11937 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004468
Annotation
PREDICTED:_inactive_dipeptidyl_peptidase_10_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.293
Sequence
CDS
ATGGTACTTATAAAAGATAAGTCGACTTCTAGAAACATCGCAGATCTTAACGTGGCCTGTCTTGGTGCTATAAATATGAAGTTCTTGTTGGTTATACCGATGTGCGTGCTGGCTGCCGGCGGTGGTCTCCTCGACGTGGATGTCGATATTTTATCCGCACCGGATAAAATCGAAGCGGATAACCTATATAGCAGTGTCATCACCGGTGACATTGACAAAGCTGTGGTCCAAACTTTGGCGCTCAACCTACTATCAAAAGGAGCCATCATCGAAGATGTCATTGCCAGGCTCATTAGAGATAAAAAGCGAAACATCTTCGACTATGCCTATAAACTATGGAACGGTGAAGGAAAGGATGTCGTCAAACAACACTTCCCTATGCAATTCAGACTGATCTTTGCCGAGAATCATGTCAAGATCATCAACAAGAGGGACAATCTCGCCATTAAACTCAGCACTGAGGTGGATCACCAAAATGACAGATCTGCATTTGGTGACGCGAACGAAAAAACCACTGACAACGTCGGCTGGAGGTTTGTCCACTTGTGGGAAAACAACAAGGTTTACTTCAAGATCGTTAGCGTTGCCCGTAACCAATTCTTGAAACTGGGTACTGATGCTGACGAAGCTGGCGACCACATCGCTTACGGTGCCGACGTGGCTAACACCTTCAGACACCAGTGGTACCTACAGCCTGCCAAGTACGAAGACGACCTTCTTTTCCTCATTTACAACCGCGAATTCAACCAGGTTCTGAAGCTCGGTCGAGTAGTCGACAACATGGGTGACCGACAAGAAATTTTGGACAGTCGAACAGCACTGTGGATGTCTGCTGATGGCCACATGGTATTGTATGCAACCTTCAACGACAGCCTCGTTCATGAGCAGAAGTTCCCCTGGTACGGCGCCGCGCTGGACACGGATGACCCTGAAAAGACATATCCAGAAATTAAGAGCGTGAGATACCCGAAGCCGGGTACCAACAACCCAACATTTACTTTAACAGTGGCGGATATCGCAGACCCTAAACATATAAGGACCAGACATTTGACGCCACCCAAAGTTATACTAGAACAAGGAGATTACTACTTCACAAGCGCGCAATGGGTGTCGTTGACTGAGGTATGCGTTGTTTGGCTCACGAGGGTCCAGAATCTGTCTGTCGTATCAGTTTGCAAGATCCCGATGTGGTTCTGTCAAGAGGTATACCGAATATCGTCAAGCAACGAAGGCTGGGTCGAATCCGCACCGGCGCCATTGTGGTCTGCAGGAGGAGGCGCACTGGTAACGCTTGCCCCAGTAAGAGACGGCCCGGCTGGTCTCTTCAGACACATCGTGAGGACTGAACACAATTCACACGGACCAAGGGCGTTACCTCTGACTCATGGTAGCTTCGACGTCGTGAAATTATTAGCGTGGGATCATGCGAACCAGCACATATATTACCTCGGCATACCAGAAGGCAAACCAGGGCAACAGCATTTATACAGGGTATCGTCTGAAGCGCCCAGACCAGGCTCACCGCAAAAACTGCCTTACTGCGTCACTTGCAATTCACAGCCTTCTCCGTCAATAAGTTTGGAGTATTACGGTAATTTGGCGTCATCAGGAGAAAGTAATTGGGACAACGATTGGGACGAAGAGCTGCCAACCACTTCACCTTCGCCCACTAAAAAGAAGAAAAGGCGACATCCAGAGGGTGTGCCGCAGAACTTGCCTTGTTTGTATCACGAGGCACACTTCAGTCCAACGGCGGCGTATTTCGTGCTGGAGTGCCTCGGTCCCGGCGTTCCGACGTCAAGCCTCCACAAGACCGCGCTTCCGGAACCAAGACTAGTCATGCACTTGGAAAACAATACGGTTGTGAAGGAGCGACTGTCGTTGATAGCGCTGCCCACACCGAGAACGTTCTCGGTTCATTTGTCCAGCGGACACCAAGCCCGGGTACGCCTGCTTCTACCACCGGGTCTTAGAGAAGACGAAGTTACCAAGTACCCTCTGGTGTTAAAAGTGCACGGAGCTCCAGGTAGCCAGCTTGTGACAGAGCGCTGGGCTGTTGACTGGGGGTCATTGGCCGCTGGTGCCGGGGCCATTGTGGCTACGGTTGACGCCAGAGGGGCTGGCGGCAGAGGCTTAGGAGCTCACCATGCTCTGCATAGAAGACTGGGGACCGTAGAGTTGCAAGATCAACTGGAAGTGGCCGAGTATCTTCGTGACTCCCTGCACTTCATAGACGCGCGCCGTGTGGCGGTGTGGGGGCGGTCGCATGGCGGTTTCCTGGCGACAATGGCGCTCGCGAGCACGCCCAGCGTCTTCCACTGCGGCATCGCAGTCACGCCTATCGTGCGCTGGCGTTACTACGCGTCAGCATACGCTGAGCGTTATATGGGCTTTCCAAACGCAACGGGCAACTACAGAGGCTACGCTGAAGCTGACGTCACCAAGCAAGCGGCGGCCTTACACGACAAGATGCTGCTGCTAGTACACGGCACAGCAGACGACAGTGTCCATATACAGCAGACTATGGCATTGGCAAGGTCATTGGCTGAACAAGGCAGCATGTTCCGGCAGCAGATTTACCCGGATGAAGGTCACAGCTTGGAAGGCGTGAAACATCACTTATACCGCACGATGTCTTCGTTCCTCGACGACTGCTTCAGGAAGCAGGTTCCTCCCGAGACCAAGGCGGGACTCAGGAACGGCGGAAACCTTGACTGA
Protein
MVLIKDKSTSRNIADLNVACLGAINMKFLLVIPMCVLAAGGGLLDVDVDILSAPDKIEADNLYSSVITGDIDKAVVQTLALNLLSKGAIIEDVIARLIRDKKRNIFDYAYKLWNGEGKDVVKQHFPMQFRLIFAENHVKIINKRDNLAIKLSTEVDHQNDRSAFGDANEKTTDNVGWRFVHLWENNKVYFKIVSVARNQFLKLGTDADEAGDHIAYGADVANTFRHQWYLQPAKYEDDLLFLIYNREFNQVLKLGRVVDNMGDRQEILDSRTALWMSADGHMVLYATFNDSLVHEQKFPWYGAALDTDDPEKTYPEIKSVRYPKPGTNNPTFTLTVADIADPKHIRTRHLTPPKVILEQGDYYFTSAQWVSLTEVCVVWLTRVQNLSVVSVCKIPMWFCQEVYRISSSNEGWVESAPAPLWSAGGGALVTLAPVRDGPAGLFRHIVRTEHNSHGPRALPLTHGSFDVVKLLAWDHANQHIYYLGIPEGKPGQQHLYRVSSEAPRPGSPQKLPYCVTCNSQPSPSISLEYYGNLASSGESNWDNDWDEELPTTSPSPTKKKKRRHPEGVPQNLPCLYHEAHFSPTAAYFVLECLGPGVPTSSLHKTALPEPRLVMHLENNTVVKERLSLIALPTPRTFSVHLSSGHQARVRLLLPPGLREDEVTKYPLVLKVHGAPGSQLVTERWAVDWGSLAAGAGAIVATVDARGAGGRGLGAHHALHRRLGTVELQDQLEVAEYLRDSLHFIDARRVAVWGRSHGGFLATMALASTPSVFHCGIAVTPIVRWRYYASAYAERYMGFPNATGNYRGYAEADVTKQAAALHDKMLLLVHGTADDSVHIQQTMALARSLAEQGSMFRQQIYPDEGHSLEGVKHHLYRTMSSFLDDCFRKQVPPETKAGLRNGGNLD
Summary
Similarity
Belongs to the peptidase S9B family.
Uniprot
A0A194Q6R3
A0A0N0PDX1
A0A1Y1KFN8
A0A067QZ10
A0A2J7Q706
A0A1B6IAV5
+ More
A0A1W4WXZ3 A0A1W4X7E1 D6WS52 N6TJZ3 A0A1B6D6U3 A0A151IP36 E2BWN1 A0A151XBV9 A0A195B8H1 F4WNQ3 A0A195FVM5 A0A0J7L9U6 A0A348G605 B0WXF5 E1ZZE7 E0VHA2 A0A0C9QQU5 A0A1I8QA09 A0A1I8QA05 A0A026WCY0 A0A3L8D6D0 A0A2A3EP60 A0A088AAR1 A0A182VME3 A0A1I8JT93 W8B5T7 A0A182Q303 A0A1B0C0S4 A0A084VMQ0 A0A0L0C4G4 A0A023F0W9 A0A224X793 A0A1I8N6D4 A0A182JBQ3 A0A336M1A4 A0A1A9WZH6 A0A182Y8J5 A0A1A9ZFA6 A0A0L7QR72 W8AJG8 A0A1B0FQM3 A0A1A9UH36 W8ARK1 A0A1J1HNU0 Q7PPP3 A0A158NVL5 B4MUJ8 W5JCT9 A0A1W4UNI7 A0A1I8N6D2 Q9VMB4 A0A1A9XT63 B4P0B5 A0A3B0KPP3 Q29JS9 B4GJ74 B4JPX9 B3N5T7 B4LVE2 B4KHT7 T1IAY1 A0A0M4ECC5 A0A0J9TH36 A0A1W4UBH9 A0A1W4UQD2 X2J9P4 B3MNY9 A0A0R3NVK4 A0A0R1DKN8 A0A0Q9XIU7 A0A0P9A5S7 A0A0M9AAU0 A0A226F1G1 A0A226F2E1
A0A1W4WXZ3 A0A1W4X7E1 D6WS52 N6TJZ3 A0A1B6D6U3 A0A151IP36 E2BWN1 A0A151XBV9 A0A195B8H1 F4WNQ3 A0A195FVM5 A0A0J7L9U6 A0A348G605 B0WXF5 E1ZZE7 E0VHA2 A0A0C9QQU5 A0A1I8QA09 A0A1I8QA05 A0A026WCY0 A0A3L8D6D0 A0A2A3EP60 A0A088AAR1 A0A182VME3 A0A1I8JT93 W8B5T7 A0A182Q303 A0A1B0C0S4 A0A084VMQ0 A0A0L0C4G4 A0A023F0W9 A0A224X793 A0A1I8N6D4 A0A182JBQ3 A0A336M1A4 A0A1A9WZH6 A0A182Y8J5 A0A1A9ZFA6 A0A0L7QR72 W8AJG8 A0A1B0FQM3 A0A1A9UH36 W8ARK1 A0A1J1HNU0 Q7PPP3 A0A158NVL5 B4MUJ8 W5JCT9 A0A1W4UNI7 A0A1I8N6D2 Q9VMB4 A0A1A9XT63 B4P0B5 A0A3B0KPP3 Q29JS9 B4GJ74 B4JPX9 B3N5T7 B4LVE2 B4KHT7 T1IAY1 A0A0M4ECC5 A0A0J9TH36 A0A1W4UBH9 A0A1W4UQD2 X2J9P4 B3MNY9 A0A0R3NVK4 A0A0R1DKN8 A0A0Q9XIU7 A0A0P9A5S7 A0A0M9AAU0 A0A226F1G1 A0A226F2E1
Pubmed
26354079
28004739
24845553
18362917
19820115
23537049
+ More
20798317 21719571 20566863 24508170 30249741 24495485 24438588 26108605 25474469 25315136 25244985 12364791 21347285 17994087 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 22936249 23185243 18057021
20798317 21719571 20566863 24508170 30249741 24495485 24438588 26108605 25474469 25315136 25244985 12364791 21347285 17994087 20920257 23761445 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 15632085 22936249 23185243 18057021
EMBL
KQ459460
KPJ00695.1
KQ460009
KPJ18468.1
GEZM01085036
GEZM01085034
+ More
JAV60299.1 KK852816 KDR15783.1 NEVH01017448 PNF24362.1 GECU01023630 GECU01014090 JAS84076.1 JAS93616.1 KQ971354 EFA07086.2 APGK01035170 APGK01035171 APGK01035172 APGK01035173 APGK01035174 APGK01035175 KB740923 ENN78213.1 GEDC01015891 JAS21407.1 KQ976892 KYN07226.1 GL451165 EFN79886.1 KQ982316 KYQ57844.1 KQ976565 KYM80540.1 GL888237 EGI64246.1 KQ981215 KYN44513.1 LBMM01000102 KMR04941.1 FX985544 BBF97878.1 DS232164 EDS36485.1 GL435343 EFN73439.1 DS235165 EEB12758.1 GBYB01006104 GBYB01006105 JAG75871.1 JAG75872.1 KK107261 EZA53922.1 QOIP01000012 RLU15804.1 KZ288210 PBC32919.1 APCN01000644 GAMC01017869 JAB88686.1 AXCN02000635 JXJN01023739 ATLV01014616 KE524975 KFB39244.1 JRES01000933 KNC27137.1 GBBI01004073 JAC14639.1 GFTR01008150 JAW08276.1 UFQT01000408 UFQT01001675 SSX24025.1 SSX31383.1 KQ414784 KOC61125.1 GAMC01017870 JAB88685.1 CCAG010002792 CCAG010002793 GAMC01017868 JAB88687.1 CVRI01000014 CRK89709.1 AAAB01008944 EAA10083.5 ADTU01002890 ADTU01002891 ADTU01002892 CH963857 EDW76193.2 ADMH02001599 ETN61886.1 AE014134 AY122228 AAF52407.3 AAM52740.1 CM000157 EDW87873.1 OUUW01000010 SPP85818.1 CH379062 EAL32882.2 CH479184 EDW37388.1 CH916372 EDV98959.1 CH954177 EDV59096.1 CH940649 EDW63321.2 CH933807 EDW13374.1 ACPB03005088 CP012523 ALC39260.1 CM002910 KMY88550.1 AHN54197.1 CH902620 EDV32176.1 KRT05034.1 KRT05035.1 KRJ97470.1 KRG03719.1 KRG03720.1 KPU73830.1 KPU73831.1 KQ435711 KOX79483.1 LNIX01000001 OXA63602.1 OXA63607.1
JAV60299.1 KK852816 KDR15783.1 NEVH01017448 PNF24362.1 GECU01023630 GECU01014090 JAS84076.1 JAS93616.1 KQ971354 EFA07086.2 APGK01035170 APGK01035171 APGK01035172 APGK01035173 APGK01035174 APGK01035175 KB740923 ENN78213.1 GEDC01015891 JAS21407.1 KQ976892 KYN07226.1 GL451165 EFN79886.1 KQ982316 KYQ57844.1 KQ976565 KYM80540.1 GL888237 EGI64246.1 KQ981215 KYN44513.1 LBMM01000102 KMR04941.1 FX985544 BBF97878.1 DS232164 EDS36485.1 GL435343 EFN73439.1 DS235165 EEB12758.1 GBYB01006104 GBYB01006105 JAG75871.1 JAG75872.1 KK107261 EZA53922.1 QOIP01000012 RLU15804.1 KZ288210 PBC32919.1 APCN01000644 GAMC01017869 JAB88686.1 AXCN02000635 JXJN01023739 ATLV01014616 KE524975 KFB39244.1 JRES01000933 KNC27137.1 GBBI01004073 JAC14639.1 GFTR01008150 JAW08276.1 UFQT01000408 UFQT01001675 SSX24025.1 SSX31383.1 KQ414784 KOC61125.1 GAMC01017870 JAB88685.1 CCAG010002792 CCAG010002793 GAMC01017868 JAB88687.1 CVRI01000014 CRK89709.1 AAAB01008944 EAA10083.5 ADTU01002890 ADTU01002891 ADTU01002892 CH963857 EDW76193.2 ADMH02001599 ETN61886.1 AE014134 AY122228 AAF52407.3 AAM52740.1 CM000157 EDW87873.1 OUUW01000010 SPP85818.1 CH379062 EAL32882.2 CH479184 EDW37388.1 CH916372 EDV98959.1 CH954177 EDV59096.1 CH940649 EDW63321.2 CH933807 EDW13374.1 ACPB03005088 CP012523 ALC39260.1 CM002910 KMY88550.1 AHN54197.1 CH902620 EDV32176.1 KRT05034.1 KRT05035.1 KRJ97470.1 KRG03719.1 KRG03720.1 KPU73830.1 KPU73831.1 KQ435711 KOX79483.1 LNIX01000001 OXA63602.1 OXA63607.1
Proteomes
UP000053268
UP000053240
UP000027135
UP000235965
UP000192223
UP000007266
+ More
UP000019118 UP000078542 UP000008237 UP000075809 UP000078540 UP000007755 UP000078541 UP000036403 UP000002320 UP000000311 UP000009046 UP000095300 UP000053097 UP000279307 UP000242457 UP000005203 UP000075903 UP000075840 UP000075886 UP000092460 UP000030765 UP000037069 UP000095301 UP000075880 UP000091820 UP000076408 UP000092445 UP000053825 UP000092444 UP000078200 UP000183832 UP000007062 UP000005205 UP000007798 UP000000673 UP000192221 UP000000803 UP000092443 UP000002282 UP000268350 UP000001819 UP000008744 UP000001070 UP000008711 UP000008792 UP000009192 UP000015103 UP000092553 UP000007801 UP000053105 UP000198287
UP000019118 UP000078542 UP000008237 UP000075809 UP000078540 UP000007755 UP000078541 UP000036403 UP000002320 UP000000311 UP000009046 UP000095300 UP000053097 UP000279307 UP000242457 UP000005203 UP000075903 UP000075840 UP000075886 UP000092460 UP000030765 UP000037069 UP000095301 UP000075880 UP000091820 UP000076408 UP000092445 UP000053825 UP000092444 UP000078200 UP000183832 UP000007062 UP000005205 UP000007798 UP000000673 UP000192221 UP000000803 UP000092443 UP000002282 UP000268350 UP000001819 UP000008744 UP000001070 UP000008711 UP000008792 UP000009192 UP000015103 UP000092553 UP000007801 UP000053105 UP000198287
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A194Q6R3
A0A0N0PDX1
A0A1Y1KFN8
A0A067QZ10
A0A2J7Q706
A0A1B6IAV5
+ More
A0A1W4WXZ3 A0A1W4X7E1 D6WS52 N6TJZ3 A0A1B6D6U3 A0A151IP36 E2BWN1 A0A151XBV9 A0A195B8H1 F4WNQ3 A0A195FVM5 A0A0J7L9U6 A0A348G605 B0WXF5 E1ZZE7 E0VHA2 A0A0C9QQU5 A0A1I8QA09 A0A1I8QA05 A0A026WCY0 A0A3L8D6D0 A0A2A3EP60 A0A088AAR1 A0A182VME3 A0A1I8JT93 W8B5T7 A0A182Q303 A0A1B0C0S4 A0A084VMQ0 A0A0L0C4G4 A0A023F0W9 A0A224X793 A0A1I8N6D4 A0A182JBQ3 A0A336M1A4 A0A1A9WZH6 A0A182Y8J5 A0A1A9ZFA6 A0A0L7QR72 W8AJG8 A0A1B0FQM3 A0A1A9UH36 W8ARK1 A0A1J1HNU0 Q7PPP3 A0A158NVL5 B4MUJ8 W5JCT9 A0A1W4UNI7 A0A1I8N6D2 Q9VMB4 A0A1A9XT63 B4P0B5 A0A3B0KPP3 Q29JS9 B4GJ74 B4JPX9 B3N5T7 B4LVE2 B4KHT7 T1IAY1 A0A0M4ECC5 A0A0J9TH36 A0A1W4UBH9 A0A1W4UQD2 X2J9P4 B3MNY9 A0A0R3NVK4 A0A0R1DKN8 A0A0Q9XIU7 A0A0P9A5S7 A0A0M9AAU0 A0A226F1G1 A0A226F2E1
A0A1W4WXZ3 A0A1W4X7E1 D6WS52 N6TJZ3 A0A1B6D6U3 A0A151IP36 E2BWN1 A0A151XBV9 A0A195B8H1 F4WNQ3 A0A195FVM5 A0A0J7L9U6 A0A348G605 B0WXF5 E1ZZE7 E0VHA2 A0A0C9QQU5 A0A1I8QA09 A0A1I8QA05 A0A026WCY0 A0A3L8D6D0 A0A2A3EP60 A0A088AAR1 A0A182VME3 A0A1I8JT93 W8B5T7 A0A182Q303 A0A1B0C0S4 A0A084VMQ0 A0A0L0C4G4 A0A023F0W9 A0A224X793 A0A1I8N6D4 A0A182JBQ3 A0A336M1A4 A0A1A9WZH6 A0A182Y8J5 A0A1A9ZFA6 A0A0L7QR72 W8AJG8 A0A1B0FQM3 A0A1A9UH36 W8ARK1 A0A1J1HNU0 Q7PPP3 A0A158NVL5 B4MUJ8 W5JCT9 A0A1W4UNI7 A0A1I8N6D2 Q9VMB4 A0A1A9XT63 B4P0B5 A0A3B0KPP3 Q29JS9 B4GJ74 B4JPX9 B3N5T7 B4LVE2 B4KHT7 T1IAY1 A0A0M4ECC5 A0A0J9TH36 A0A1W4UBH9 A0A1W4UQD2 X2J9P4 B3MNY9 A0A0R3NVK4 A0A0R1DKN8 A0A0Q9XIU7 A0A0P9A5S7 A0A0M9AAU0 A0A226F1G1 A0A226F2E1
PDB
1Z68
E-value=1.816e-72,
Score=696
Ontologies
GO
Topology
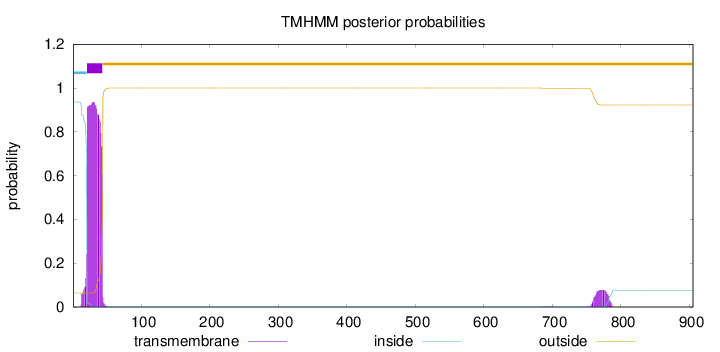
Length:
905
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.99388
Exp number, first 60 AAs:
21.25191
Total prob of N-in:
0.93570
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 905
Population Genetic Test Statistics
Pi
262.448311
Theta
171.87225
Tajima's D
1.449518
CLR
0.782601
CSRT
0.778961051947403
Interpretation
Uncertain
