Gene
KWMTBOMO11933
Annotation
hypothetical_protein_[Piscirickettsia_salmonis]
Location in the cell
Mitochondrial Reliability : 1.399 Nuclear Reliability : 1.188
Sequence
CDS
ATGTATTGTTCTCCTAACCCAAAAAAAACTGATGAAAGTGTGGGCGGCAACACGGGCCGCGAAATATCTACGGCACCGTTTAAGGTGCCAACCGTAGCAGCAAAAGCTGTAAGATGCGAGAGCGCACTCTCGATGCGGTCGGGAAGTGCTGCTTCCGTGAGGAGTAGCGCCTCGAGTGACCGAGGGTCGCTCTCGGTAGATGAAATGAATAGTACGATTGTAGGAGGAAGAGCACTCCGCAAACGTAAAACGGCTCCGCGAGCGGCGGCTTCCCGCCCTTCTGACTCCTCGGAGGGGGAACCCGCACCGCCCGCGAAATTCCTCGCCTTAGAGGAAACCACGGCACCAGTCGGTGGTCGCCAGGTGGCAACGGACTCTGCCAGTGCAGAGGTCGAGCGACTCGTCCGCTCCCTCACCGAGGTGGGGTTCGAAGACTCCACAAAACTATTTAGGGTCGCCTCGGATATTGTATCCAGTGTGGCAGCCAAATCGGGCAGGGGTTTTAAGGGCATATACCTGCATGCTCTTAAGTGCTTGGCTGCCACAATTGAAGACTACCTAAACATAGTTACTCACAAGACGAGCGGCGAGAACGAGGAATGCGTGCGACTGCAGAGAGACCAGCTCAAGGCACAAAACGCCGAGCTGCAAAATAATAATCTTGAGCTACGGGCGGAGATCGCGACCCTACACATGCGGGTGGACGAACTCATGGCGGAGATACGTAGCCCGGCTCCACCCGATGTCGAGGAGGTAGCTGAGCTAAAGCGGCGGATCGCCATTTTAGAGGCGCGCAGTTCGCCAGTGGAGCGTGCAAGACCACTGCTCGCACACGAGCGGAGAGCTGGCACCCAGGTCAAGCCCCAGTCTGCCAAACAGAATAAGCAGCCTGCCAAGCACGATCAAACCCAGCCGACCACCCTGGCCAAGCTTCATGTAGCTAAGCAAGCCAAGGCCCAGCAGCCGGTCCCTACCAAGCCACAGCCTGCCGTAGCGCGTACTGGAAGGGACCGAACGAAGAAGAAAGGCGTGAATGCTGCCAAAGTGATGCAGTACCCTCAGCCCTGCCCGCTGCCTCCGGTCCCATCCAGCATGGACGTAGCGTGGACGACAGTGGTAAAGCGGGGACAAAGAAAGACGAAGGAAGGACAAAGAAGCCCTCGGTCTGTCCAGGCAGTTGCCGCTCCCCACGTAGAGGGTCCAGCGGTGAAGATACGGAAGAACCGTCGAGGAGGAAGATCCGGCAAGACACGCGCTCCGCGGTCGGCTGCAGTTGTTTTAGAATTGCTGCCGGCCGCTAAAGACAAGGGTATCACTTACGAAGAAGTGATGGCCCGGGCGCGTGGTGGCGTGGATATTGACGCCATCGGAGTGGAAGGCGGACTGAAGGTTCGACACACGGCTAACGGCGCTCGTTTGCTCGAATGTCCCGGTGCTGACTCAGGAGCGGTTGCGGACCGACTTATGGCCCGACTCCGCGAGATCTTGCCTGATCCGGAGGTGGTGAGGATCGACAGGCCTGTCAGAATGGCGGAGATCAAGGTGACGGGCCTGGACGAGTGCGCCATGAAAGAGGAGGTGGCTGCCGCTATCGCATTGCAGGGGAACTGTGCTCTTGCAAACGTCAAGGTGGGGGAGCTTCGGAGTACATATTCCGGAACTCGCTCTGTATGGGCGCGGTGCCCCGTACAGACAGCCAGCCCGTACTGGCTACGCCTGCACAGGGAAAGCCCGCTGACACGCCCGGTAGGCTACGCGTGGGGTGGGTCATTGCCCACGTGCAGTTGCAGGAAGCTCGACCATGGCGATGCCTTCGATGTTTCGGCACCGGGCACGGCCTCGCCAAGTGCTCGTCAGCTGTTGACCGCAGCGGACTATGTTTCCGGTGCGGCCAATCTGGGCACAAGGCCGCTTCCTGCACTGCCGCCATGCCACACTGTGTATTGTGCGACGCGGAGAAGCGGCGGGCCAACCACCGGGTTGGTGGCCCAGCATGTCATTTTACCTCTTTCTCATCCAAGAGGAGGAAGGGCGAATTTGAACCACTCCGCCAGGGCGCAGGATCTCCTCGTCCACACCATGGTGGAGTGGTCAATCGATGTTGCCATAGCCGCAGAGCCGTACTTCGTCCCTGCGGACCAGGATGCCTGGATCGGGGACGTCGACGGCTCCGTGGCTATTACTATCAGGCAGGCCGCGGCGTTACCCCCCCTGGTGATGGTGGCCAGGGGATCCGGTTATGTCGCGGCCAGGGTCGACGAAACCGTCGTGATCGGGGTATACTTCTCTCCGAATAGACATCTCGCCGAGTTCGAGCAGTTCCTGGGCGGGCTTGAGGTGTTGGTTCACCGCCTCGGGTCCCGCCCGGTTATCTTAGCGGGAGACCTCAACGCAAAATGGGTGACTTGGGGTTCCCCCCGCTCGGACTCACGCGGCAGGATGCTCTCGGAGTGGGCGATCGCGGCTGGTCTCTGTCTTTTGAACAGAGGTTCGGTCGCGACTTGCGTGCGATGGAACGGCGAATCTCACGTGGACGTGACATTCGCATCTCCGTCCATCGCACGCCGCACCCGTGGCTGGCGCGTTCTTGAGGGGGCGGAGACGCTCTCGGACCATCGATACGTCCGATTTGAGCTCTCAGCCTCCTCTCGATCCGCGTCGGCTATGAACGCCCGCGGTGAGGAGGAGCTCCCGCGAAGTGCTCCTCGGTCATTCCCCAGGTGGGCACTGAAGCGCCTGAATAAGGAGCTCCTGGTGGAGGCAGCCACGGTGGCTGCGTGGGCACCCGTGCCTACTAGCCCCGTGGACGTGGAGTCAGAGGCTGACTGGTTCCGGGGAACGATGGGTCGTATTTGTGACTCAGCGATGCCCCGGGTAGGCAGCAATGCTCCGCGTGGCGGTGCGTACTGGTGGTCGCCCGAAATCGCGCAACTCCGCGAGGAGTGCGTGCGGGCGCGCCGCCGGAGCGCTCGCCATCGCCGCCGCCGCCTTCGCGATGAGGACTTCGCGGAGGTAGCGGACCGGTTGCGCGCCGACTGCCGCCAAAAGCAGGAAGCGTTGCGACTGGCCATCGGTGAGGCCAAATCTCAGGGCATGCAGACGCTCCTGGAGATGCTCGATCAGGATCCCTGGGGGCGCCCGTACCAAACGGTCCGCAAGAAATTGCGGCCGTGGGCGCTCCCAGTGACGGAGCGTCTCCAGCCTCAGCAGCTGCGGGAGATTGTCTCCGGGCTGTTCCCGCAGATGGAGAGGGACTTTGAGCCTCCCTCCATGGGCGCGCCGCCTCGCAGTGACGTGAGCGGTATTGCTGAGGTGGTCCCTCCGAGCATTTTGGAGGAAGAGATTCATGCGGCCGTGTTGAGGATGCGGAGAAAAGACGCGGCCCCCGGCCCTGACGGTGTTCACGGCCGGGTCTGGGATTTGGCCTTCGGTGCCCTTGGGGACCGGCTCGTGCGGCTCTTTGAAGCCTGCTTCGAGTCGGGACGGTTTCCTAAGCAGTGGAAGACGGGCAGACTCGTCCTTCTGAGGAAGGAGGGGCGCCCCGCAGACTCACCTGCGGGGTACCGCCCCATCGTGTTGCTGGACGAGGCGGGAAAATTGTTGGAGCGAGTTATCGCCGCCCGCATCGTCCGGCACCTGACCGGGACGGGTCCCGATCTGTCGGAGGAACAGTATGGCTTCAGAGAGGGCCGCTCTACTATCGACGCGATTATGCGCGTGCGTGCCCTCTCTGACGAAGCTGTCGCCCGGGGTGGGGTGGCCTTGGCGGTGTCCCTGGATATCGCCAACGCTTTTAACACCTTGCCCTGGTCAGTGATCGCGGGGGCGCTCGAATATCATGGCGTCCCCGCGTACCTCCGCCGACTGATCGGGTCCTATCTCGAGGACAGATCGGTCGTGTGCACCGGACACGGTGGTGCGGTGCTCCGCTTTCCGGTCGAGCGCGGTGTTCCACAGGGGTCGGTCCTTGGCCCTCTCCTGTGGAATATCGGTTACGACTGGGCTCTGAGGAGCGCCCTGCACGGCCCTCTCCCGGGCCTAAGCGTTGTGTGTTACGCGGATGACACGTTGGTCGTGGCCCGGGGGAGGAACTACCAAGAGTCTGCTCGTCTTGCCTGCGCAGGGGTGGCATTCGTCGTCGGTAAGATCCGAAGGCTGGGCCTCGACGTGGCCCTCGAGAAATCCCAGGCCCTGCTGTTTCACAGGGCTGGGAGAGAGCCGCCACCCGGGGCCCACCTGGTGATTGGAGGCGTCCGCGTCGAGGTCGGGGTGACTGGGTTACGGTACCTCGGTCTCGAATTGGACGGTCGTTGGAACTTCCGCGCTCATTTCGAGAAGTTGGGCCCCCGACTGATGGCGATGGCTGGATCGCTGAGTCGGCTCTTGCCAAACGTCGGAGGGCCTGACACGGTGGTGCGCCGCTTCTACGTGGGGGTGGTGCGGTCTATTGCACTATACGGAGCCCCCGTGTGGTGCCGCGCCTTGACTCGCAGAAACGTCGTGGCGCTGCGACGCCCGCAGCGTGCAATCGCAGTCAGGGCGATTCGTGGATACCGCACCGTCTCCTTCGAGGCGGCGTGCGTTCTCGCCGGGACGCCTCCCTGGGACCTGGAGGCGGAGGCGCTCGCTGCCGATTATGCGTGGCGTTGTGACCTTCGCGACAGGGGGGAACCACGTCCCGGAGAAGGAGCAATTAGAGCGCGGAGGCTCCAATCTCGGCGGTCTGTGTTGGAGGCGTGGTCTCGCCGCTTGGCGGACCCGTCGGCCGGTCTCCGGACCGTCGAGGCGGTGCGCCCGGTCCTCGCGAAATGGGCGAACCGTGACCAAGGGCGTCTCACCTTCCGGTTGACGCAAATGCTTACCGGCCATGGGTGTTTCGGCCGGTACCTGCATCGCGTTGCCCGGAGGGAACCGACGATGGAGTGTCACCACTGTGACTGCGACGAGGACACGGTCGAGCATACGCTCGCATATTGCCCCGCATGGGTGGAGCAACGCCGTGTCCTTGTCTCGCAAATAGGACCGGACTTGTCGCTGCCGACTGTCGTGGCTACGATGCTCGGCAGCGATGAGTCCTGGAAGGCGATGCTCGACTTCTGCGAGTGCACCATCTCGCAGAAGGAGGCGGCGGAACGAGAGAGGGAGAGCTCATCCCTCTCCGCACCTATCCGTCGCCGCCGAGCCGGGGGCCGCAGAAGGGCTTTTGTCCGACTTCGGCCCCTGTAA
Protein
MYCSPNPKKTDESVGGNTGREISTAPFKVPTVAAKAVRCESALSMRSGSAASVRSSASSDRGSLSVDEMNSTIVGGRALRKRKTAPRAAASRPSDSSEGEPAPPAKFLALEETTAPVGGRQVATDSASAEVERLVRSLTEVGFEDSTKLFRVASDIVSSVAAKSGRGFKGIYLHALKCLAATIEDYLNIVTHKTSGENEECVRLQRDQLKAQNAELQNNNLELRAEIATLHMRVDELMAEIRSPAPPDVEEVAELKRRIAILEARSSPVERARPLLAHERRAGTQVKPQSAKQNKQPAKHDQTQPTTLAKLHVAKQAKAQQPVPTKPQPAVARTGRDRTKKKGVNAAKVMQYPQPCPLPPVPSSMDVAWTTVVKRGQRKTKEGQRSPRSVQAVAAPHVEGPAVKIRKNRRGGRSGKTRAPRSAAVVLELLPAAKDKGITYEEVMARARGGVDIDAIGVEGGLKVRHTANGARLLECPGADSGAVADRLMARLREILPDPEVVRIDRPVRMAEIKVTGLDECAMKEEVAAAIALQGNCALANVKVGELRSTYSGTRSVWARCPVQTASPYWLRLHRESPLTRPVGYAWGGSLPTCSCRKLDHGDAFDVSAPGTASPSARQLLTAADYVSGAANLGTRPLPALPPCHTVYCATRRSGGPTTGLVAQHVILPLSHPRGGRANLNHSARAQDLLVHTMVEWSIDVAIAAEPYFVPADQDAWIGDVDGSVAITIRQAAALPPLVMVARGSGYVAARVDETVVIGVYFSPNRHLAEFEQFLGGLEVLVHRLGSRPVILAGDLNAKWVTWGSPRSDSRGRMLSEWAIAAGLCLLNRGSVATCVRWNGESHVDVTFASPSIARRTRGWRVLEGAETLSDHRYVRFELSASSRSASAMNARGEEELPRSAPRSFPRWALKRLNKELLVEAATVAAWAPVPTSPVDVESEADWFRGTMGRICDSAMPRVGSNAPRGGAYWWSPEIAQLREECVRARRRSARHRRRRLRDEDFAEVADRLRADCRQKQEALRLAIGEAKSQGMQTLLEMLDQDPWGRPYQTVRKKLRPWALPVTERLQPQQLREIVSGLFPQMERDFEPPSMGAPPRSDVSGIAEVVPPSILEEEIHAAVLRMRRKDAAPGPDGVHGRVWDLAFGALGDRLVRLFEACFESGRFPKQWKTGRLVLLRKEGRPADSPAGYRPIVLLDEAGKLLERVIAARIVRHLTGTGPDLSEEQYGFREGRSTIDAIMRVRALSDEAVARGGVALAVSLDIANAFNTLPWSVIAGALEYHGVPAYLRRLIGSYLEDRSVVCTGHGGAVLRFPVERGVPQGSVLGPLLWNIGYDWALRSALHGPLPGLSVVCYADDTLVVARGRNYQESARLACAGVAFVVGKIRRLGLDVALEKSQALLFHRAGREPPPGAHLVIGGVRVEVGVTGLRYLGLELDGRWNFRAHFEKLGPRLMAMAGSLSRLLPNVGGPDTVVRRFYVGVVRSIALYGAPVWCRALTRRNVVALRRPQRAIAVRAIRGYRTVSFEAACVLAGTPPWDLEAEALAADYAWRCDLRDRGEPRPGEGAIRARRLQSRRSVLEAWSRRLADPSAGLRTVEAVRPVLAKWANRDQGRLTFRLTQMLTGHGCFGRYLHRVARREPTMECHHCDCDEDTVEHTLAYCPAWVEQRRVLVSQIGPDLSLPTVVATMLGSDESWKAMLDFCECTISQKEAAERERESSSLSAPIRRRRAGGRRRAFVRLRPL
Summary
EMBL
Proteomes
Interpro
Gene 3D
ProteinModelPortal
PDB
1WDU
E-value=2.94486e-09,
Score=154
Ontologies
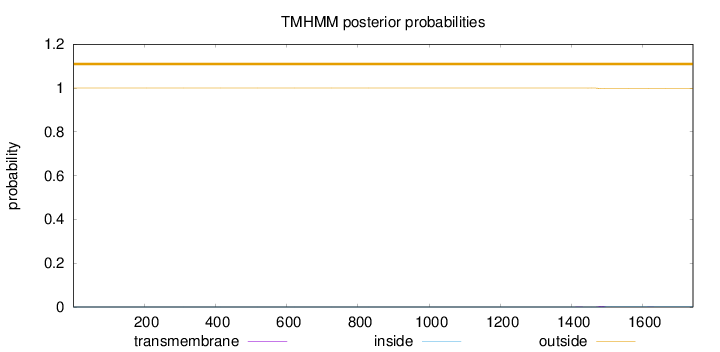
Topology
Length:
1741
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06153
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00006
outside
1 - 1741
Population Genetic Test Statistics
Pi
12.945894
Theta
14.11249
Tajima's D
0
CLR
0.171598
CSRT
0.374231288435578
Interpretation
Uncertain
