Gene
KWMTBOMO11927
Pre Gene Modal
BGIBMGA004461
Annotation
PREDICTED:_inactive_dipeptidyl_peptidase_10_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.565
Sequence
CDS
ATGGTTAAGCTGGTTGTGGGTGGTCCGAACCAGCGCAACTGGCGCGGCATCCTGATCGCATTACTGGTCATCGTGGCTGTCCTCGCCCTCATAGTCACTTCCGTGGTGCTTCTGACTCCTCCTGACGAAGGCCCGAGGGTCAAAGGCAGGAGGTTCCGCATCCAAGACTTACTCAACAATGAATTGGAACCGATCAGATGGAACGGGACGTGGATATCGGGTGACGAGTTTTTGTACCGCGACGCATGGGGCGGTATCAGTTTGATGTTCGCAGCCAATCAGACCTCGAAGACGCTGATGCCAAATTCAACATTTCAAACCATTAGCAGAGAGAAATGCGATGTATAA
Protein
MVKLVVGGPNQRNWRGILIALLVIVAVLALIVTSVVLLTPPDEGPRVKGRRFRIQDLLNNELEPIRWNGTWISGDEFLYRDAWGGISLMFAANQTSKTLMPNSTFQTISREKCDV
Summary
Similarity
Belongs to the peptidase S9B family.
Uniprot
A0A0N0PDX1
A0A3S2NVQ9
D6WS52
A0A067QZ10
A0A336M6M1
A0A336MLU4
+ More
A0A1B6D6U3 A0A0M9AAU0 A0A1W4WXZ3 A0A1W4X7E1 A0A0C9QQU5 A0A0C9RGD7 A0A1I8N6D2 A0A1B0FQM3 A0A182JBQ3 A0A1I8N6D4 B3N5T7 A0A158NVL5 A0A1A9XT63 N6TJZ3 A0A151XBV9 B4Q4Y7 A0A195B8H1 A0A0Q9XIU7 B4KHT7 A0A1A9WZH6 B4HXU5 X2J9P4 A0A1I8QA05 Q9VMB4 A0A0J9TH36 W8AJG8 B4LVE2 A0A1I8QA09 W8ARK1 A0A088AAR1 A0A151IP36 W8B5T7 B4JPX9 A0A3B0KPP3 A0A0P9A5S7 B3MNY9 A0A1W4UQD2 A0A0R3NVK4 A0A1W4UBH9 A0A1W4UNI7 Q29JS9 B4GJ74 F4WNQ3 A0A0K8VTI2 B4P0B5 A0A0R1DKN8 A0A2A3EP60 E1ZZE7 E2BWN1 A0A195FVM5 B4MUJ8 A0A0M4ECC5 A0A026WCY0 A0A3L8D6D0 A0A023F0W9 A0A1J1HNU0 A0A224X793 A0A067QPP4 A0A1D2MHC3 T1IAY1 A0A2R7X781 A0A1B0C0S4 A0A226F1G1 D6WQR4 A0A1A9UH36 A0A1A9ZFA6 A0A1A9YR64 N6TI84 A0A226F2E1 B4R662 Q9VX90 M9PHX1 Q8IR04 B4Q246 B3NWA7 U4U5H3 A0A147BUP0 B3N0E7 A0A1D2N010 A0A1B6E895 A0A0M4EVB7
A0A1B6D6U3 A0A0M9AAU0 A0A1W4WXZ3 A0A1W4X7E1 A0A0C9QQU5 A0A0C9RGD7 A0A1I8N6D2 A0A1B0FQM3 A0A182JBQ3 A0A1I8N6D4 B3N5T7 A0A158NVL5 A0A1A9XT63 N6TJZ3 A0A151XBV9 B4Q4Y7 A0A195B8H1 A0A0Q9XIU7 B4KHT7 A0A1A9WZH6 B4HXU5 X2J9P4 A0A1I8QA05 Q9VMB4 A0A0J9TH36 W8AJG8 B4LVE2 A0A1I8QA09 W8ARK1 A0A088AAR1 A0A151IP36 W8B5T7 B4JPX9 A0A3B0KPP3 A0A0P9A5S7 B3MNY9 A0A1W4UQD2 A0A0R3NVK4 A0A1W4UBH9 A0A1W4UNI7 Q29JS9 B4GJ74 F4WNQ3 A0A0K8VTI2 B4P0B5 A0A0R1DKN8 A0A2A3EP60 E1ZZE7 E2BWN1 A0A195FVM5 B4MUJ8 A0A0M4ECC5 A0A026WCY0 A0A3L8D6D0 A0A023F0W9 A0A1J1HNU0 A0A224X793 A0A067QPP4 A0A1D2MHC3 T1IAY1 A0A2R7X781 A0A1B0C0S4 A0A226F1G1 D6WQR4 A0A1A9UH36 A0A1A9ZFA6 A0A1A9YR64 N6TI84 A0A226F2E1 B4R662 Q9VX90 M9PHX1 Q8IR04 B4Q246 B3NWA7 U4U5H3 A0A147BUP0 B3N0E7 A0A1D2N010 A0A1B6E895 A0A0M4EVB7
Pubmed
EMBL
KQ460009
KPJ18468.1
RSAL01000053
RVE50035.1
KQ971354
EFA07086.2
+ More
KK852816 KDR15783.1 UFQT01000408 SSX24027.1 UFQT01001675 SSX31384.1 GEDC01015891 JAS21407.1 KQ435711 KOX79483.1 GBYB01006104 GBYB01006105 JAG75871.1 JAG75872.1 GBYB01006106 JAG75873.1 CCAG010002792 CCAG010002793 CH954177 EDV59096.1 ADTU01002890 ADTU01002891 ADTU01002892 APGK01035170 APGK01035171 APGK01035172 APGK01035173 APGK01035174 APGK01035175 KB740923 ENN78213.1 KQ982316 KYQ57844.1 CM000361 EDX03979.1 KQ976565 KYM80540.1 CH933807 KRG03719.1 KRG03720.1 EDW13374.1 CH480818 EDW51875.1 AE014134 AHN54197.1 AY122228 AAF52407.3 AAM52740.1 CM002910 KMY88550.1 GAMC01017870 JAB88685.1 CH940649 EDW63321.2 GAMC01017868 JAB88687.1 KQ976892 KYN07226.1 GAMC01017869 JAB88686.1 CH916372 EDV98959.1 OUUW01000010 SPP85818.1 CH902620 KPU73830.1 KPU73831.1 EDV32176.1 CH379062 KRT05034.1 KRT05035.1 EAL32882.2 CH479184 EDW37388.1 GL888237 EGI64246.1 GDHF01010122 JAI42192.1 CM000157 EDW87873.1 KRJ97470.1 KZ288210 PBC32919.1 GL435343 EFN73439.1 GL451165 EFN79886.1 KQ981215 KYN44513.1 CH963857 EDW76193.2 CP012523 ALC39260.1 KK107261 EZA53922.1 QOIP01000012 RLU15804.1 GBBI01004073 JAC14639.1 CVRI01000014 CRK89709.1 GFTR01008150 JAW08276.1 KK853083 KDR11620.1 LJIJ01001239 ODM92407.1 ACPB03005088 KK858101 PTY27632.1 JXJN01023739 LNIX01000001 OXA63602.1 EFA06037.2 APGK01025705 APGK01025706 KB740600 ENN80119.1 OXA63607.1 CM000366 EDX18166.1 AE014298 AAF48688.3 AGB95474.1 AAN09430.3 CM000162 EDX02554.2 CH954180 EDV46446.2 KB631982 ERL87588.1 GEGO01001359 JAR94045.1 CH902640 EDV38351.2 LJIJ01000344 ODM98541.1 GEDC01003140 JAS34158.1 CP012528 ALC49185.1
KK852816 KDR15783.1 UFQT01000408 SSX24027.1 UFQT01001675 SSX31384.1 GEDC01015891 JAS21407.1 KQ435711 KOX79483.1 GBYB01006104 GBYB01006105 JAG75871.1 JAG75872.1 GBYB01006106 JAG75873.1 CCAG010002792 CCAG010002793 CH954177 EDV59096.1 ADTU01002890 ADTU01002891 ADTU01002892 APGK01035170 APGK01035171 APGK01035172 APGK01035173 APGK01035174 APGK01035175 KB740923 ENN78213.1 KQ982316 KYQ57844.1 CM000361 EDX03979.1 KQ976565 KYM80540.1 CH933807 KRG03719.1 KRG03720.1 EDW13374.1 CH480818 EDW51875.1 AE014134 AHN54197.1 AY122228 AAF52407.3 AAM52740.1 CM002910 KMY88550.1 GAMC01017870 JAB88685.1 CH940649 EDW63321.2 GAMC01017868 JAB88687.1 KQ976892 KYN07226.1 GAMC01017869 JAB88686.1 CH916372 EDV98959.1 OUUW01000010 SPP85818.1 CH902620 KPU73830.1 KPU73831.1 EDV32176.1 CH379062 KRT05034.1 KRT05035.1 EAL32882.2 CH479184 EDW37388.1 GL888237 EGI64246.1 GDHF01010122 JAI42192.1 CM000157 EDW87873.1 KRJ97470.1 KZ288210 PBC32919.1 GL435343 EFN73439.1 GL451165 EFN79886.1 KQ981215 KYN44513.1 CH963857 EDW76193.2 CP012523 ALC39260.1 KK107261 EZA53922.1 QOIP01000012 RLU15804.1 GBBI01004073 JAC14639.1 CVRI01000014 CRK89709.1 GFTR01008150 JAW08276.1 KK853083 KDR11620.1 LJIJ01001239 ODM92407.1 ACPB03005088 KK858101 PTY27632.1 JXJN01023739 LNIX01000001 OXA63602.1 EFA06037.2 APGK01025705 APGK01025706 KB740600 ENN80119.1 OXA63607.1 CM000366 EDX18166.1 AE014298 AAF48688.3 AGB95474.1 AAN09430.3 CM000162 EDX02554.2 CH954180 EDV46446.2 KB631982 ERL87588.1 GEGO01001359 JAR94045.1 CH902640 EDV38351.2 LJIJ01000344 ODM98541.1 GEDC01003140 JAS34158.1 CP012528 ALC49185.1
Proteomes
UP000053240
UP000283053
UP000007266
UP000027135
UP000053105
UP000192223
+ More
UP000095301 UP000092444 UP000075880 UP000008711 UP000005205 UP000092443 UP000019118 UP000075809 UP000000304 UP000078540 UP000009192 UP000091820 UP000001292 UP000000803 UP000095300 UP000008792 UP000005203 UP000078542 UP000001070 UP000268350 UP000007801 UP000192221 UP000001819 UP000008744 UP000007755 UP000002282 UP000242457 UP000000311 UP000008237 UP000078541 UP000007798 UP000092553 UP000053097 UP000279307 UP000183832 UP000094527 UP000015103 UP000092460 UP000198287 UP000078200 UP000092445 UP000030742
UP000095301 UP000092444 UP000075880 UP000008711 UP000005205 UP000092443 UP000019118 UP000075809 UP000000304 UP000078540 UP000009192 UP000091820 UP000001292 UP000000803 UP000095300 UP000008792 UP000005203 UP000078542 UP000001070 UP000268350 UP000007801 UP000192221 UP000001819 UP000008744 UP000007755 UP000002282 UP000242457 UP000000311 UP000008237 UP000078541 UP000007798 UP000092553 UP000053097 UP000279307 UP000183832 UP000094527 UP000015103 UP000092460 UP000198287 UP000078200 UP000092445 UP000030742
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A0N0PDX1
A0A3S2NVQ9
D6WS52
A0A067QZ10
A0A336M6M1
A0A336MLU4
+ More
A0A1B6D6U3 A0A0M9AAU0 A0A1W4WXZ3 A0A1W4X7E1 A0A0C9QQU5 A0A0C9RGD7 A0A1I8N6D2 A0A1B0FQM3 A0A182JBQ3 A0A1I8N6D4 B3N5T7 A0A158NVL5 A0A1A9XT63 N6TJZ3 A0A151XBV9 B4Q4Y7 A0A195B8H1 A0A0Q9XIU7 B4KHT7 A0A1A9WZH6 B4HXU5 X2J9P4 A0A1I8QA05 Q9VMB4 A0A0J9TH36 W8AJG8 B4LVE2 A0A1I8QA09 W8ARK1 A0A088AAR1 A0A151IP36 W8B5T7 B4JPX9 A0A3B0KPP3 A0A0P9A5S7 B3MNY9 A0A1W4UQD2 A0A0R3NVK4 A0A1W4UBH9 A0A1W4UNI7 Q29JS9 B4GJ74 F4WNQ3 A0A0K8VTI2 B4P0B5 A0A0R1DKN8 A0A2A3EP60 E1ZZE7 E2BWN1 A0A195FVM5 B4MUJ8 A0A0M4ECC5 A0A026WCY0 A0A3L8D6D0 A0A023F0W9 A0A1J1HNU0 A0A224X793 A0A067QPP4 A0A1D2MHC3 T1IAY1 A0A2R7X781 A0A1B0C0S4 A0A226F1G1 D6WQR4 A0A1A9UH36 A0A1A9ZFA6 A0A1A9YR64 N6TI84 A0A226F2E1 B4R662 Q9VX90 M9PHX1 Q8IR04 B4Q246 B3NWA7 U4U5H3 A0A147BUP0 B3N0E7 A0A1D2N010 A0A1B6E895 A0A0M4EVB7
A0A1B6D6U3 A0A0M9AAU0 A0A1W4WXZ3 A0A1W4X7E1 A0A0C9QQU5 A0A0C9RGD7 A0A1I8N6D2 A0A1B0FQM3 A0A182JBQ3 A0A1I8N6D4 B3N5T7 A0A158NVL5 A0A1A9XT63 N6TJZ3 A0A151XBV9 B4Q4Y7 A0A195B8H1 A0A0Q9XIU7 B4KHT7 A0A1A9WZH6 B4HXU5 X2J9P4 A0A1I8QA05 Q9VMB4 A0A0J9TH36 W8AJG8 B4LVE2 A0A1I8QA09 W8ARK1 A0A088AAR1 A0A151IP36 W8B5T7 B4JPX9 A0A3B0KPP3 A0A0P9A5S7 B3MNY9 A0A1W4UQD2 A0A0R3NVK4 A0A1W4UBH9 A0A1W4UNI7 Q29JS9 B4GJ74 F4WNQ3 A0A0K8VTI2 B4P0B5 A0A0R1DKN8 A0A2A3EP60 E1ZZE7 E2BWN1 A0A195FVM5 B4MUJ8 A0A0M4ECC5 A0A026WCY0 A0A3L8D6D0 A0A023F0W9 A0A1J1HNU0 A0A224X793 A0A067QPP4 A0A1D2MHC3 T1IAY1 A0A2R7X781 A0A1B0C0S4 A0A226F1G1 D6WQR4 A0A1A9UH36 A0A1A9ZFA6 A0A1A9YR64 N6TI84 A0A226F2E1 B4R662 Q9VX90 M9PHX1 Q8IR04 B4Q246 B3NWA7 U4U5H3 A0A147BUP0 B3N0E7 A0A1D2N010 A0A1B6E895 A0A0M4EVB7
Ontologies
GO
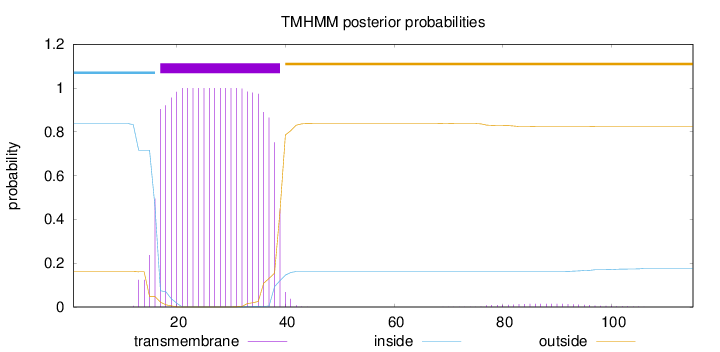
Topology
Length:
115
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
23.02071
Exp number, first 60 AAs:
22.74509
Total prob of N-in:
0.83777
POSSIBLE N-term signal
sequence
inside
1 - 16
TMhelix
17 - 39
outside
40 - 115
Population Genetic Test Statistics
Pi
317.381254
Theta
176.417141
Tajima's D
3.041186
CLR
0.150897
CSRT
0.980550972451377
Interpretation
Uncertain
