Gene
KWMTBOMO11890
Pre Gene Modal
BGIBMGA004453
Annotation
PREDICTED:_developmental_protein_eyes_absent_[Bombyx_mori]
Full name
Eyes absent homolog
Location in the cell
Nuclear Reliability : 1.92
Sequence
CDS
ATGAATGACGACAGCACCGGTGGCGTAAAAAGCGAGGTGCGGAGTCCGGGGCTTTGCGAGGCAGACGTGGCGCTGTACGGGGAAACGCTTCCATACGACCAGACCGCGCTGCCGTACCAATACTACAATAGTATGCAGCAGTATGGTAGCGGCAGCGCGGCTTCGTCGTACGTCAGCTCTGGTCTTTACAACCAGCCTTACGCTGCATATCCTCCACCGCACAACACGAACAACAGATCGTCATGCAAAGCGACGCCTACATACCTCAGTGCGTACGGGATGAGTGGAGTGGGAAGTGTGCCCGCCACAGGATTCGGCACTCAACCCTCGCCGTACGCATACTCTTCATACAACGGAGGACTATCGCAAACCTTCACTCCTACACAACAAGATTATAGTAATTATGCAGCCGGCTACGCGGAACACAATGTTGCTCAGTACAGTGGCTATTACGCAGCACCCAGCTATTCACCTTATGTCAGTTCGCCAAGCAGTAGTGGGAGCGCCGGTCACACTTCATACCATCTCGGAGGAGCATTGTCAGATTCACCATCATCAATGTTGCCATCAATTAATGATACGAGTCCATTATCTCCGATTAAAGCTGAAGTACACGCGGCCAGCAGAAGATGTAGAGAGAATTCTGGGGAAAGCACAGCTAGTAGGTCTCGAGGTCGAGGACGCAGGAACACCTCATCATCGCCAGCCCAGCACGTCCCAGAGCCAGCCACAGACAGAGTCTTCATATGGGATTTGGACGAGACTATAATTATATTCCATTCGTTGCTCACTGGAACTTATGCTACTAAGTACAATAAGGATACGCAACAGATAGTGCAATTAGGATTTAGGATGGAAGAAATGATATTCAGCTTGGCTGATACACATTTCTTTTTCAATGATGTTGAGGATTGCGACCAGGAGCACATAGACGCGGTATCGGCCGATGATAATGGCCAGGATCTCTCTGCGTACAACTTCACAACGGATGGGTTTCATGCGGGCGCGGCGGGAGGCGCTCTGTGTGCGCCTGGGGTGCGAGGCGGAGTCGACTGGATGAGGAAACTAGCCTTCCGGTACAGGAAGATCAAAGACACCTACAATAATTACAGGAATAGTGTTGGAGGGCTCCTAGGGCCAGCGAAGAGAGAACAGTGGTTACAGTTGAGAGCAGAACTTGAACAGGCCACGGATAACTGGTTAACATTAGCCTGCAAGTGTCTGAATATGATTAATTCTCGAGAGAACTGCGTGAATGTTTTAGTGACGACGACTCAGCTCGTCCCGGCTTTGGCTAAAGTCCTGCTCTTCGGTTTAGGGGGCGTGTTTCCTATCGAAAATATATACTCCGCCACGAAAACGGGTAAGGAGACTTGCTTTGAGAAAATTAAGCAGCGCTTCGGCGAGCGGTGCACGTACGTCGTCATAGGGGACGGCCAGGACGAGGAGGCCGCCGCCAAGGCCAAGAACTACCCTTTCTGGAGGGTGTCGAGCCACTCCGACATCGCGGCGCTGTACAACGCGCTCGACATGGGCTTCTTATGA
Protein
MNDDSTGGVKSEVRSPGLCEADVALYGETLPYDQTALPYQYYNSMQQYGSGSAASSYVSSGLYNQPYAAYPPPHNTNNRSSCKATPTYLSAYGMSGVGSVPATGFGTQPSPYAYSSYNGGLSQTFTPTQQDYSNYAAGYAEHNVAQYSGYYAAPSYSPYVSSPSSSGSAGHTSYHLGGALSDSPSSMLPSINDTSPLSPIKAEVHAASRRCRENSGESTASRSRGRGRRNTSSSPAQHVPEPATDRVFIWDLDETIIIFHSLLTGTYATKYNKDTQQIVQLGFRMEEMIFSLADTHFFFNDVEDCDQEHIDAVSADDNGQDLSAYNFTTDGFHAGAAGGALCAPGVRGGVDWMRKLAFRYRKIKDTYNNYRNSVGGLLGPAKREQWLQLRAELEQATDNWLTLACKCLNMINSRENCVNVLVTTTQLVPALAKVLLFGLGGVFPIENIYSATKTGKETCFEKIKQRFGERCTYVVIGDGQDEEAAAKAKNYPFWRVSSHSDIAALYNALDMGFL
Summary
Catalytic Activity
H2O + O-phospho-L-tyrosyl-[protein] = L-tyrosyl-[protein] + phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the HAD-like hydrolase superfamily. EYA family.
Uniprot
EC Number
3.1.3.48
Pubmed
EMBL
NWSH01000046
PCG80242.1
KZ150066
PZC74135.1
BABH01021921
KQ971354
+ More
KYB26324.1 KB632263 ERL90918.1 GECZ01003578 JAS66191.1 GEZM01093997 JAV56019.1 GECL01001733 JAP04391.1 GBBI01002735 JAC15977.1 ACPB03005128 GBHO01041401 GBHO01022696 JAG02203.1 JAG20908.1 GBHO01022698 GBHO01022695 GBRD01007973 GBRD01007972 GDHC01011693 JAG20906.1 JAG20909.1 JAG57848.1 JAQ06936.1 KP168452 AJV21307.1
KYB26324.1 KB632263 ERL90918.1 GECZ01003578 JAS66191.1 GEZM01093997 JAV56019.1 GECL01001733 JAP04391.1 GBBI01002735 JAC15977.1 ACPB03005128 GBHO01041401 GBHO01022696 JAG02203.1 JAG20908.1 GBHO01022698 GBHO01022695 GBRD01007973 GBRD01007972 GDHC01011693 JAG20906.1 JAG20909.1 JAG57848.1 JAQ06936.1 KP168452 AJV21307.1
Proteomes
PRIDE
Interpro
SUPFAM
SSF56784
SSF56784
Gene 3D
CDD
ProteinModelPortal
PDB
3HB1
E-value=3.18149e-111,
Score=1028
Ontologies
KEGG
GO
PANTHER
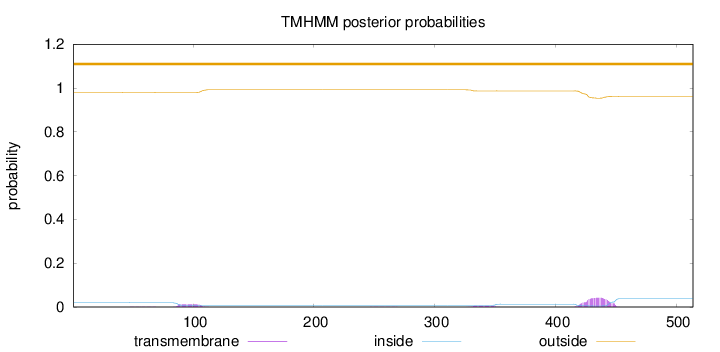
Topology
Length:
514
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.29131
Exp number, first 60 AAs:
0.00315
Total prob of N-in:
0.02001
outside
1 - 514
Population Genetic Test Statistics
Pi
200.780659
Theta
147.100266
Tajima's D
0.605487
CLR
0.297661
CSRT
0.548172591370432
Interpretation
Uncertain
