Gene
KWMTBOMO11883
Pre Gene Modal
BGIBMGA004407
Annotation
PREDICTED:_serine/threonine-protein_phosphatase_2A_regulatory_subunit_B''_subunit_gamma-like_[Papilio_xuthus]
Location in the cell
Cytoplasmic Reliability : 2.108 Nuclear Reliability : 1.917
Sequence
CDS
ATGTCTGATAAAGAAAAAGAAGTGAAAAATAAGGGCAGTGGAACCGACATTAGTGCTTCTGGGGACGCCAAGCTGCCGAACGAACAGGACACCAAATTGAATAATATTTTGAAAAACAGACTCAAAAAATACATTAAAAAACTAGACGATACTGGAACGAGCATAACAAAACAAGAAAGTGAAACAATTGAAGTGGAGACATTCAAGGAACTTTATCAACCCAAATTGGACAATGATGAATCTTACAAAAGCATACCTAAATTTTACTACAAATTACCCCGTAATGATGATGTCCTAGCACAGAAACTACGAGAAGAAACTCGAGCACAATTTCTGCAGAAGAAGAGCAAGGAGCTACTCGATAATGCTGAGTTGAAACATCTCTGGACCCTGTTAGAGAAGACCAACGGGAATTACAGCAGCTCCAACAATGATGAGTTGATCATAGACTATTTTCAATTTAAGAAGATCAGAGAAGAAGCTGGACCTAAATATAGACCGTATTTCACAGCAGAAGTTTTTGGACGTCTGCAAGCAGCCGAAGGCAGTGTTGGCAGGATTCGTGCTGTCTCTCTATTCAACTATGTTATGAGACGAGTGTGGCTGCAGCAGACGAGAATCGGACTATCTCTCTATGATGTTACGGGACAAGGCTATCTCACTGAGCAAGATTTGGAGAGTTACATAGCAGAGTTAGTCCCGTCTCTGGCAGCTCTAGACGGTCTGGACTCATCATTCACCTCGTTCTACGTATGCACGGCAGCGAGGAAGTTCCTTTTCTTCCTCGACCCACTACGGATCGGACGTGTAAGGATCAGCGACGTCCTATCCTGTTCTTTCTTAGATGATTTACTTGAGCTCCGCGAAGAAGACCTACCGATGGAGTTGCAAGAGCAGAACTGGTTCAGTGCGGCCTCGGCGCTACGCGTCTACGGGCAGTACTTGAACTTGGACAGAGACCACAACGGGATGCTGAGTATAAACGAACTGGCCGGCTACGGCTCCGGCACCCTGACCCAGGCCTTCCTCCAGCGCGTCTTCCAGCAGTGCCTCACGTACGACGGCGAGATGGACTACAAGACGTACCTGGACCTGGTCCTCGCCCTCGAGAACCGGAAGCAGCCCGCCGCGCTCGCCTATCTGTTCAGAGTGTTGGACATCAATTCCGTGGGCTACCTCGACGCTTTCACCTTGAATTATTTCTTTAAGGCGATACAGAAACAGATGATAGCCCACGGAGCCGAGCCGGTCAATTTCGATGATGTGAAAGACGAGATATTCGACATGATCCGACCTAAAGACCCGACCAAGATCACACTTCAAGACCTGGTGAGGAGTGGTCACGGGCACACCGCAGTCTCCATACTGCTGGAGCTGCACGGCTTCTGGGCTTACGAGAACAGGGAGGCTCTGGCAGCCGCCGATCACAACACTTGA
Protein
MSDKEKEVKNKGSGTDISASGDAKLPNEQDTKLNNILKNRLKKYIKKLDDTGTSITKQESETIEVETFKELYQPKLDNDESYKSIPKFYYKLPRNDDVLAQKLREETRAQFLQKKSKELLDNAELKHLWTLLEKTNGNYSSSNNDELIIDYFQFKKIREEAGPKYRPYFTAEVFGRLQAAEGSVGRIRAVSLFNYVMRRVWLQQTRIGLSLYDVTGQGYLTEQDLESYIAELVPSLAALDGLDSSFTSFYVCTAARKFLFFLDPLRIGRVRISDVLSCSFLDDLLELREEDLPMELQEQNWFSAASALRVYGQYLNLDRDHNGMLSINELAGYGSGTLTQAFLQRVFQQCLTYDGEMDYKTYLDLVLALENRKQPAALAYLFRVLDINSVGYLDAFTLNYFFKAIQKQMIAHGAEPVNFDDVKDEIFDMIRPKDPTKITLQDLVRSGHGHTAVSILLELHGFWAYENREALAAADHNT
Summary
Uniprot
H9J4G7
A0A2A4JBK4
A0A2H1WCM2
A0A3S2NH87
A0A194Q6T0
A0A151JUA9
+ More
A0A151I5W2 F4WQ39 A0A195ECB1 A0A026VTW9 A0A151WY83 A0A088AKV4 E9IVY9 A0A2A3EQ06 A0A310SF10 E2C5X1 A0A158NWX3 A0A151IHK7 A0A232EVV4 K7IVQ5 A0A0C9R6H0 A0A1W4WM29 A0A1A8QLM1 A0A1A8N2S4 A0A1A8GA33 A0A1W4WC00 A0A0T6B6C7 A0A1W4WN15 A0A1A8L071 A0A1A8ECX3 A0A3Q2DK31 A0A091VHQ1 A0A1A7XC04 A0A0S7H4X1 A0A091RJT3 A0A3B3YJC9 A0A087Y6Q6 A0A3B3W0V1 A0A1A8ALT4 A0A3L8SZM7 A0A091VA88 M4A4K4 A0A3P9NUE0 A0A182WR69 A0A091W2Y8 A0A1A7Z462 A0A147AC67 A0A067QG16 A0A091KQ41 A0A0A0APB2 A0A091TDQ2 A0A093NUM4 A0A091SQX2 A0A093PYR4 K7FKL5 F1N899 A0A091KFD7 A0A091RUY1 A0A093HIK3 A0A091HIR1 A0A3Q1IAN5 A0A3Q1BL98 A0A3P8RTV7 W5MS39 A0A182V0K7 A0A182TZ59 A0A3Q3KP34 A0A091F370 A0A3S2PBA2 A0A091PPZ8 M3XCC3 A0A3Q1CKV4 A0A3B3DFK7 A0A3P8RTW7 A0A2U3VWK2 I3MD47 A0A384CC12 F6UYZ0 A0A3Q0EIS5 A0A3Q7Y025 A0A151NLA0 A0A0F8CDN0 R0LTB0 M7B129 A0A0B8RVK9 A0A287A1B6 A0A2Y9QYR7 A0A2Y9DL05 A0A1V4KFC1 A0A091P3P3 A0A3Q7TYE4 A0A2I4CCV6 U3ISV4 U3JU09 G5B115 A0A384CD71 G1MAT2 E2RNM2 G3T4I2
A0A151I5W2 F4WQ39 A0A195ECB1 A0A026VTW9 A0A151WY83 A0A088AKV4 E9IVY9 A0A2A3EQ06 A0A310SF10 E2C5X1 A0A158NWX3 A0A151IHK7 A0A232EVV4 K7IVQ5 A0A0C9R6H0 A0A1W4WM29 A0A1A8QLM1 A0A1A8N2S4 A0A1A8GA33 A0A1W4WC00 A0A0T6B6C7 A0A1W4WN15 A0A1A8L071 A0A1A8ECX3 A0A3Q2DK31 A0A091VHQ1 A0A1A7XC04 A0A0S7H4X1 A0A091RJT3 A0A3B3YJC9 A0A087Y6Q6 A0A3B3W0V1 A0A1A8ALT4 A0A3L8SZM7 A0A091VA88 M4A4K4 A0A3P9NUE0 A0A182WR69 A0A091W2Y8 A0A1A7Z462 A0A147AC67 A0A067QG16 A0A091KQ41 A0A0A0APB2 A0A091TDQ2 A0A093NUM4 A0A091SQX2 A0A093PYR4 K7FKL5 F1N899 A0A091KFD7 A0A091RUY1 A0A093HIK3 A0A091HIR1 A0A3Q1IAN5 A0A3Q1BL98 A0A3P8RTV7 W5MS39 A0A182V0K7 A0A182TZ59 A0A3Q3KP34 A0A091F370 A0A3S2PBA2 A0A091PPZ8 M3XCC3 A0A3Q1CKV4 A0A3B3DFK7 A0A3P8RTW7 A0A2U3VWK2 I3MD47 A0A384CC12 F6UYZ0 A0A3Q0EIS5 A0A3Q7Y025 A0A151NLA0 A0A0F8CDN0 R0LTB0 M7B129 A0A0B8RVK9 A0A287A1B6 A0A2Y9QYR7 A0A2Y9DL05 A0A1V4KFC1 A0A091P3P3 A0A3Q7TYE4 A0A2I4CCV6 U3ISV4 U3JU09 G5B115 A0A384CD71 G1MAT2 E2RNM2 G3T4I2
Pubmed
EMBL
BABH01021894
NWSH01002109
PCG69146.1
ODYU01007766
SOQ50818.1
RSAL01000039
+ More
RVE51002.1 KQ459460 KPJ00710.1 KQ981799 KYN35689.1 KQ976434 KYM87203.1 GL888262 EGI63662.1 KQ979074 KYN22860.1 KK107921 EZA47243.1 KQ982651 KYQ52839.1 GL766449 EFZ15273.1 KZ288195 PBC33787.1 KQ765323 OAD53978.1 GL452863 EFN76662.1 ADTU01002984 KQ977586 KYN01680.1 NNAY01001950 OXU22473.1 GBYB01002361 GBYB01002363 JAG72128.1 JAG72130.1 HAEH01012129 HAEI01014774 SBR94009.1 HAEF01021997 HAEG01006854 SBR63156.1 HAEB01020707 HAEC01001347 SBQ67234.1 LJIG01009774 KRT82407.1 HAED01018320 HAEE01017244 SBR37294.1 HADZ01009188 HAEA01015298 SBQ43778.1 KK733992 KFR01993.1 HADW01014236 SBP15636.1 GBYX01445353 JAO36079.1 KK818601 KFQ39209.1 AYCK01001343 HADY01017153 HAEJ01015575 SBP55638.1 QUSF01000004 RLW10209.1 KK421149 KFQ86593.1 KL410344 KFQ96183.1 HADX01015375 SBP37607.1 GCES01010242 JAR76081.1 KK853541 KDR06877.1 KK537708 KFP29904.1 KL872281 KGL94890.1 KK446689 KFQ71855.1 KL224798 KFW63852.1 KK482797 KFQ61009.1 KL671375 KFW81908.1 AGCU01049778 AGCU01049779 AGCU01049780 AGCU01049781 AGCU01049782 AGCU01049783 AADN05000303 KK502300 KFP22451.1 KK934204 KFQ46430.1 KL206140 KFV79275.1 KL536880 KFO94712.1 AHAT01003900 KK719449 KFO63711.1 CM012458 RVE57006.1 KK643413 KFP97382.1 AANG04004459 AGTP01026094 AGTP01026095 AGTP01026096 AGTP01026097 AGTP01026098 AGTP01026099 AGTP01026100 AKHW03002792 KYO37265.1 KQ042255 KKF16965.1 KB742730 EOB05010.1 KB548684 EMP30749.1 GBZA01000507 JAG69261.1 AEMK02000056 DQIR01032834 DQIR01209816 HCZ88309.1 LSYS01003385 OPJ83170.1 KK667498 KFQ02235.1 ADON01038359 AGTO01010443 JH167915 GEBF01003135 EHB02976.1 JAO00498.1 ACTA01051676 ACTA01059676 AAEX03005671
RVE51002.1 KQ459460 KPJ00710.1 KQ981799 KYN35689.1 KQ976434 KYM87203.1 GL888262 EGI63662.1 KQ979074 KYN22860.1 KK107921 EZA47243.1 KQ982651 KYQ52839.1 GL766449 EFZ15273.1 KZ288195 PBC33787.1 KQ765323 OAD53978.1 GL452863 EFN76662.1 ADTU01002984 KQ977586 KYN01680.1 NNAY01001950 OXU22473.1 GBYB01002361 GBYB01002363 JAG72128.1 JAG72130.1 HAEH01012129 HAEI01014774 SBR94009.1 HAEF01021997 HAEG01006854 SBR63156.1 HAEB01020707 HAEC01001347 SBQ67234.1 LJIG01009774 KRT82407.1 HAED01018320 HAEE01017244 SBR37294.1 HADZ01009188 HAEA01015298 SBQ43778.1 KK733992 KFR01993.1 HADW01014236 SBP15636.1 GBYX01445353 JAO36079.1 KK818601 KFQ39209.1 AYCK01001343 HADY01017153 HAEJ01015575 SBP55638.1 QUSF01000004 RLW10209.1 KK421149 KFQ86593.1 KL410344 KFQ96183.1 HADX01015375 SBP37607.1 GCES01010242 JAR76081.1 KK853541 KDR06877.1 KK537708 KFP29904.1 KL872281 KGL94890.1 KK446689 KFQ71855.1 KL224798 KFW63852.1 KK482797 KFQ61009.1 KL671375 KFW81908.1 AGCU01049778 AGCU01049779 AGCU01049780 AGCU01049781 AGCU01049782 AGCU01049783 AADN05000303 KK502300 KFP22451.1 KK934204 KFQ46430.1 KL206140 KFV79275.1 KL536880 KFO94712.1 AHAT01003900 KK719449 KFO63711.1 CM012458 RVE57006.1 KK643413 KFP97382.1 AANG04004459 AGTP01026094 AGTP01026095 AGTP01026096 AGTP01026097 AGTP01026098 AGTP01026099 AGTP01026100 AKHW03002792 KYO37265.1 KQ042255 KKF16965.1 KB742730 EOB05010.1 KB548684 EMP30749.1 GBZA01000507 JAG69261.1 AEMK02000056 DQIR01032834 DQIR01209816 HCZ88309.1 LSYS01003385 OPJ83170.1 KK667498 KFQ02235.1 ADON01038359 AGTO01010443 JH167915 GEBF01003135 EHB02976.1 JAO00498.1 ACTA01051676 ACTA01059676 AAEX03005671
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000078541
UP000078540
+ More
UP000007755 UP000078492 UP000053097 UP000075809 UP000005203 UP000242457 UP000008237 UP000005205 UP000078542 UP000215335 UP000002358 UP000192223 UP000265020 UP000053605 UP000261480 UP000028760 UP000261500 UP000276834 UP000002852 UP000242638 UP000076407 UP000053283 UP000265000 UP000027135 UP000053858 UP000054081 UP000053258 UP000007267 UP000000539 UP000053119 UP000053584 UP000265040 UP000257160 UP000265080 UP000018468 UP000075903 UP000075902 UP000261600 UP000052976 UP000011712 UP000261560 UP000245340 UP000005215 UP000261680 UP000002281 UP000189704 UP000286642 UP000050525 UP000031443 UP000008227 UP000248480 UP000190648 UP000286640 UP000192220 UP000016666 UP000016665 UP000006813 UP000008912 UP000002254 UP000007646
UP000007755 UP000078492 UP000053097 UP000075809 UP000005203 UP000242457 UP000008237 UP000005205 UP000078542 UP000215335 UP000002358 UP000192223 UP000265020 UP000053605 UP000261480 UP000028760 UP000261500 UP000276834 UP000002852 UP000242638 UP000076407 UP000053283 UP000265000 UP000027135 UP000053858 UP000054081 UP000053258 UP000007267 UP000000539 UP000053119 UP000053584 UP000265040 UP000257160 UP000265080 UP000018468 UP000075903 UP000075902 UP000261600 UP000052976 UP000011712 UP000261560 UP000245340 UP000005215 UP000261680 UP000002281 UP000189704 UP000286642 UP000050525 UP000031443 UP000008227 UP000248480 UP000190648 UP000286640 UP000192220 UP000016666 UP000016665 UP000006813 UP000008912 UP000002254 UP000007646
PRIDE
Pfam
PF17958 EF-hand_13
Interpro
SUPFAM
SSF47473
SSF47473
ProteinModelPortal
H9J4G7
A0A2A4JBK4
A0A2H1WCM2
A0A3S2NH87
A0A194Q6T0
A0A151JUA9
+ More
A0A151I5W2 F4WQ39 A0A195ECB1 A0A026VTW9 A0A151WY83 A0A088AKV4 E9IVY9 A0A2A3EQ06 A0A310SF10 E2C5X1 A0A158NWX3 A0A151IHK7 A0A232EVV4 K7IVQ5 A0A0C9R6H0 A0A1W4WM29 A0A1A8QLM1 A0A1A8N2S4 A0A1A8GA33 A0A1W4WC00 A0A0T6B6C7 A0A1W4WN15 A0A1A8L071 A0A1A8ECX3 A0A3Q2DK31 A0A091VHQ1 A0A1A7XC04 A0A0S7H4X1 A0A091RJT3 A0A3B3YJC9 A0A087Y6Q6 A0A3B3W0V1 A0A1A8ALT4 A0A3L8SZM7 A0A091VA88 M4A4K4 A0A3P9NUE0 A0A182WR69 A0A091W2Y8 A0A1A7Z462 A0A147AC67 A0A067QG16 A0A091KQ41 A0A0A0APB2 A0A091TDQ2 A0A093NUM4 A0A091SQX2 A0A093PYR4 K7FKL5 F1N899 A0A091KFD7 A0A091RUY1 A0A093HIK3 A0A091HIR1 A0A3Q1IAN5 A0A3Q1BL98 A0A3P8RTV7 W5MS39 A0A182V0K7 A0A182TZ59 A0A3Q3KP34 A0A091F370 A0A3S2PBA2 A0A091PPZ8 M3XCC3 A0A3Q1CKV4 A0A3B3DFK7 A0A3P8RTW7 A0A2U3VWK2 I3MD47 A0A384CC12 F6UYZ0 A0A3Q0EIS5 A0A3Q7Y025 A0A151NLA0 A0A0F8CDN0 R0LTB0 M7B129 A0A0B8RVK9 A0A287A1B6 A0A2Y9QYR7 A0A2Y9DL05 A0A1V4KFC1 A0A091P3P3 A0A3Q7TYE4 A0A2I4CCV6 U3ISV4 U3JU09 G5B115 A0A384CD71 G1MAT2 E2RNM2 G3T4I2
A0A151I5W2 F4WQ39 A0A195ECB1 A0A026VTW9 A0A151WY83 A0A088AKV4 E9IVY9 A0A2A3EQ06 A0A310SF10 E2C5X1 A0A158NWX3 A0A151IHK7 A0A232EVV4 K7IVQ5 A0A0C9R6H0 A0A1W4WM29 A0A1A8QLM1 A0A1A8N2S4 A0A1A8GA33 A0A1W4WC00 A0A0T6B6C7 A0A1W4WN15 A0A1A8L071 A0A1A8ECX3 A0A3Q2DK31 A0A091VHQ1 A0A1A7XC04 A0A0S7H4X1 A0A091RJT3 A0A3B3YJC9 A0A087Y6Q6 A0A3B3W0V1 A0A1A8ALT4 A0A3L8SZM7 A0A091VA88 M4A4K4 A0A3P9NUE0 A0A182WR69 A0A091W2Y8 A0A1A7Z462 A0A147AC67 A0A067QG16 A0A091KQ41 A0A0A0APB2 A0A091TDQ2 A0A093NUM4 A0A091SQX2 A0A093PYR4 K7FKL5 F1N899 A0A091KFD7 A0A091RUY1 A0A093HIK3 A0A091HIR1 A0A3Q1IAN5 A0A3Q1BL98 A0A3P8RTV7 W5MS39 A0A182V0K7 A0A182TZ59 A0A3Q3KP34 A0A091F370 A0A3S2PBA2 A0A091PPZ8 M3XCC3 A0A3Q1CKV4 A0A3B3DFK7 A0A3P8RTW7 A0A2U3VWK2 I3MD47 A0A384CC12 F6UYZ0 A0A3Q0EIS5 A0A3Q7Y025 A0A151NLA0 A0A0F8CDN0 R0LTB0 M7B129 A0A0B8RVK9 A0A287A1B6 A0A2Y9QYR7 A0A2Y9DL05 A0A1V4KFC1 A0A091P3P3 A0A3Q7TYE4 A0A2I4CCV6 U3ISV4 U3JU09 G5B115 A0A384CD71 G1MAT2 E2RNM2 G3T4I2
PDB
4I5N
E-value=1.68859e-20,
Score=246
Ontologies
GO
PANTHER
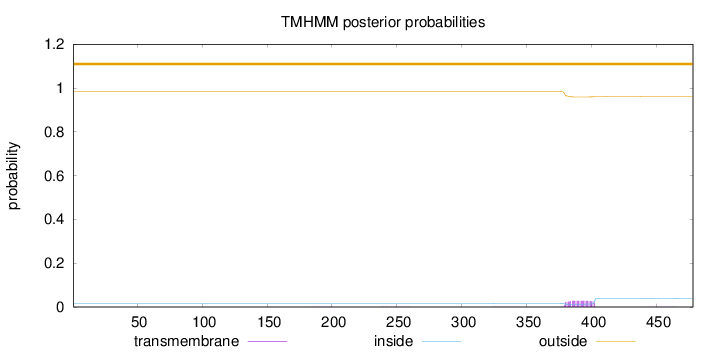
Topology
Length:
478
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.64823
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01600
outside
1 - 478
Population Genetic Test Statistics
Pi
257.792771
Theta
182.862139
Tajima's D
1.386632
CLR
0.487184
CSRT
0.760061996900155
Interpretation
Uncertain
