Gene
KWMTBOMO11882 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004447
Annotation
PREDICTED:_5-oxoprolinase_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 1.513
Sequence
CDS
ATGGGCAAAAGATTCCAGTTTGCAATAGACCGAGGAGGTACTTTTACGGATGTGTATGCACGATGCCCTAATGGAAAGGTGCGTGTCATGAAGCTGTTGTCTGTTGATCCTCAGAACTATTCCGATGCTCCCAGGGAAGCAATAAGAAGGATATTACAAGAAGAAACTGGCAATGCATTAGACAAATATGGCAAAGTCAACTCTGCCCTAATAGAATCTATTCGCATGGGTACCACAGTCGCCACAAATGCTTTGCTGGAAAGGAAAGGAGCTGCAATGGCTCTGGTCATTAACAAGGGCTTCAAAGATTTACTTTACATCGGAAACCAGGCGAGACCGAATATATTTCAACTGAACATCAAACGTCCAGAGGTCTTGTATAAAGAGGTGGTAGAGGTAGATTGTAGAGTGATACCAGCGCTAGAAGACCGATGTCAGTTGGACAAGGCTGGGAAAAATTGGAAGGAAGTTTATGGAACCACCGGTGAAAAGATGCTAGTTATCAAAGACATAGACGAAGATGCAGTCAGATTAGACTTGCAGGCGTTGAAAGAAAAGGGGATAGAGAGTATTGCTGTAGTCTTAGCGCATAGTTACACATACCACGACCACGAGATAAGAGTGGGGAAGATAGCCGAAAGCTTAGGTTTCACTCAAGTATCATTGTCGCATGCTGTAACGTCCATGGTGCGCATGGTGCCGCGAGGGTTCACGGCTTCAGCTGACGCGTACCTGACTCCTCACATCCGGGAGTACGTGAGGAGTTTCTCGGACGGCTTCACCGATTCTCTGAAGGACGCCAACGTGCTGTTTATGCAATCTGACGGTGGTTTGACTCCTATGAATTTATTCAACGGCTCGCGCGCCATCCTCTCTGGACCAGCTGGAGGCGTGATTGGTTACGCTATGACCTCGTACCAGAAAGAAACCGGCCTGCCGGTCATAGGTTTCGATATGGGCGGCACCTCGACGGACGTGTCGCGGTACGCGGGCTCCCTGGAGCACGTCCACGAGGCCACCACGGCCGGCGTCACGATACAAGCGCCACAGCTAGATATAAACACAGTGGCAGCGGGCGGCGGGTCCATGCTGTTCTTCCGGTCGGGCCTGTTCGTGGCGGGGCCGGAGTCCGCGGGCGCCGAGCCGGGCCCCGCTTGCTATCGGAAGGGCGGTCCGCTCGCCGTCACTGACGCTAATCTGTTGCTAGGTCGTCTGCTGCCTGAATACTTCCCGAAGATCTTCGGTCCCGATGAGAACGAGCCCTTGGACCGAGCCGCGACAGTGACGGCCTTCGAGAAAATGACAGAAGAAATTAATGCGTTCTTGAAACAAGATGGCGGTAACGAGATGACAACAGAAGAAGTAGCAATGGGTTTTATCAATGTTGCCAACGAAGCCATGTGTCGTCCTATTAGGGCGCTGACGACGGCGCGCGGGCATGACGCGGCGCAGCACGCGCTGGCGTGCTTCGGCGGCGCGGGCGGCCAGCACGCCTGCTCCGTGGCGCGGCGTCTGGGCATCTCCACTGTGCTCATACATAAATACGCAGGTATCCTGTCGGCATACGGTATGGCCCTGGCCCACATAGTGCACGAGGCGCAGGAGCCCAGCGCGTGCGTCTACAGCCCAGAGAACTACCAGCAGCTCGACCGCAAGCTGGATGTGCTATCTGCTGTTTGTAAGGATAAGCTGTTGGCTCAGGGTATTCCAGCGTCCCGGATAGTTTTAGAGCCGTACTTACATCTCCGGTACGCGGGGACGGACTGCGCGCTCATGGTGTCCCCCCTGCCCGGGGGCTCCGCCACGGCTTGCAGCCACGGGGACTTCTATACGGCCTTCGTGAACAGATACAAAAACGAGTTCGGTTTCACGTTGTCCGAGCGAGACGTGCTCGTGGACGACGTCCGCGTGCGCGGCGTGGGACTCAGCGCCGCGCACGAGCACGAGGGGGTGCCCAGCGGGGAAGGCGCGCCGCCCCCCGCCGAGAAGACGGTGAAGGTGTACTTCGAAGGCGGCTACCAGGATACTAAAATATATTTATTGGAGAAGCTATTGGCGGAGCAAGTGGTCCCGGGACCAGCTATTATAATGGATAGCTTGTCGACAATACTTGTCGAGCCTGATTGTCGAGCAGAAATAACAAAGTACGGAGACGTCAAAATCACTATTGGGTCTGGGCAAGCGAAGAAAGTGACCACCGCCTTGGACTCCATACAGCTCAGCATATTCTCGCATCGTTTCATGTCTATAGCTGAACAGATGGGCAGAGTACTACAAAGAACATCTATATCGGTGAACATCAAAGAGCGATTGGACTTCTCGTGCGCGCTGTTCGGTCCCACCGGTGGGCTGGTCTCGAACGCTCCTCACATCCCCGTACATTTGGGCGCTATGCAGGAGGCGGTTCAGTATCAGATGAAAGTACGCGGCGATACCCTGCGGCCCGGCGACGTGCTGTTGGCCAACCATCCCAAAGCCGGAGGTTCGCATCTGCCCGACCTCACAGTCATAACGCCGGTCTTCCACGGGAGCGACCCTCGTCCGATATTCTTCGTGGCGTCCCGCGGGCACCACGCCGACATCGGCGGCCTCACGCCGGGCTCCATGCCGCCGCACTCTGTCAGCCTCACGCAGGAGGGCGCCGCCTTCAAGTCCTTCCTGCTCGTCGACAAGGGCGCTTTCAACGAGAAACTGCTTGTCGAAGAACTGATGAAACCCGGGAACGAGCCCGGCTGTTCCGGGACCAGGAACCTGGCTGACAACCTCTCCGATCTGAAGGCGCAGGTCGCGGCCAATCAAAGGGGCATACAGCTAGTGGGGGAGTTGATAGAGGAATACGGCCTCGAAGTGGTACAGGCTTACATGGAGCACATACAGAAGAACGCCGAGCTGGCCGTGAGGGACATGCTCAAGGAGATAGCGAGGAGCGCTATTGCGGAGACAGGCTCGGCCGTGCTGTCGGCTGTCGAATACATGGACAACGGCACGCCGATCAGCCTCACCGTGACCCTGGATAAAGACTCCGGGAGCGCCCTGTGTGACTTCACCGGCACAGGAGTTGAGGTGTGGGGCAACCTGAACGCTCCCCGCGCGATCACCCTGTCGGCCCTCATCTATTGTCTGCGTTGCATGGTGGGCCACGACGTCCCCCTCAACCAGGGTTGCCTCACTCCGGTGAGCGTGGTGATACCCCCGGGCTGCATCTTGGACCCGTCGGAGGGGGCGGCCGTGGTGGGCGGCAACGTGCTCACCTCGCAGCGCATCGTCGACGTCGTGCTCAAGGCTTTTCAGGTGTGTGCGGCGTCGCAGGGGTGCATGAACAACCTGACTCTAGGGGAGGCCGACTGGGGGTACTACGAGACCGTGGCGGGTGGCAGCGGAGCTGGTCCGGGGTGGGCGGGGCAGGGCGGCGTCCACACGCACATGACCAACACTCGCATCACGGACGTCGAGATCGTGGAGCGTCGGTACCCCATGCTGGTCACCGGGTTCAGCCTCAGAGCCGGCTCGGGTGGTAGAGGGCGCTGGGCGGGCGGCGAGGGCGTGCGGCGCGAGCTGGTGCTACGGCGCGCCGTGCGTCTCTCTGTGCTCAGCGAGCGCCGCGCGCTGCCCCCCTACGGGCTCCGCGGCGGCGAGGCTGGGGCCCGTGGACTCAACCTACTAGAGCGAACAGACGGCAGGATCATCAATCTCGGTGGAAAGGCGTCTGTTGACGCTTATCCAGGGGACAAATACATAATGAATTCGCCCGGAGGTGGGGGGTATGGTCCCCCGGCGGAAAGCTCGAGTGACGAACAAACAGACAAACAATACAGCGCGTTTATTGAGCGGGGTAGCGTGTACGAGTATAGGAGTGCTCAGGAATCGGTTTAA
Protein
MGKRFQFAIDRGGTFTDVYARCPNGKVRVMKLLSVDPQNYSDAPREAIRRILQEETGNALDKYGKVNSALIESIRMGTTVATNALLERKGAAMALVINKGFKDLLYIGNQARPNIFQLNIKRPEVLYKEVVEVDCRVIPALEDRCQLDKAGKNWKEVYGTTGEKMLVIKDIDEDAVRLDLQALKEKGIESIAVVLAHSYTYHDHEIRVGKIAESLGFTQVSLSHAVTSMVRMVPRGFTASADAYLTPHIREYVRSFSDGFTDSLKDANVLFMQSDGGLTPMNLFNGSRAILSGPAGGVIGYAMTSYQKETGLPVIGFDMGGTSTDVSRYAGSLEHVHEATTAGVTIQAPQLDINTVAAGGGSMLFFRSGLFVAGPESAGAEPGPACYRKGGPLAVTDANLLLGRLLPEYFPKIFGPDENEPLDRAATVTAFEKMTEEINAFLKQDGGNEMTTEEVAMGFINVANEAMCRPIRALTTARGHDAAQHALACFGGAGGQHACSVARRLGISTVLIHKYAGILSAYGMALAHIVHEAQEPSACVYSPENYQQLDRKLDVLSAVCKDKLLAQGIPASRIVLEPYLHLRYAGTDCALMVSPLPGGSATACSHGDFYTAFVNRYKNEFGFTLSERDVLVDDVRVRGVGLSAAHEHEGVPSGEGAPPPAEKTVKVYFEGGYQDTKIYLLEKLLAEQVVPGPAIIMDSLSTILVEPDCRAEITKYGDVKITIGSGQAKKVTTALDSIQLSIFSHRFMSIAEQMGRVLQRTSISVNIKERLDFSCALFGPTGGLVSNAPHIPVHLGAMQEAVQYQMKVRGDTLRPGDVLLANHPKAGGSHLPDLTVITPVFHGSDPRPIFFVASRGHHADIGGLTPGSMPPHSVSLTQEGAAFKSFLLVDKGAFNEKLLVEELMKPGNEPGCSGTRNLADNLSDLKAQVAANQRGIQLVGELIEEYGLEVVQAYMEHIQKNAELAVRDMLKEIARSAIAETGSAVLSAVEYMDNGTPISLTVTLDKDSGSALCDFTGTGVEVWGNLNAPRAITLSALIYCLRCMVGHDVPLNQGCLTPVSVVIPPGCILDPSEGAAVVGGNVLTSQRIVDVVLKAFQVCAASQGCMNNLTLGEADWGYYETVAGGSGAGPGWAGQGGVHTHMTNTRITDVEIVERRYPMLVTGFSLRAGSGGRGRWAGGEGVRRELVLRRAVRLSVLSERRALPPYGLRGGEAGARGLNLLERTDGRIINLGGKASVDAYPGDKYIMNSPGGGGYGPPAESSSDEQTDKQYSAFIERGSVYEYRSAQESV
Summary
Uniprot
H9J4K7
A0A067RWH5
A0A2J7QAU6
A0A087ZYN6
A0A151XCJ6
A0A3L8DPT6
+ More
A0A1Y1NFB5 A0A310SQ07 E2BGJ9 A0A232FMW6 A0A195EXZ2 E2B0R6 A0A158NLJ1 A0A1B6E0J6 A0A151IMT7 A0A195B1G5 F4WW16 A0A195E6E3 A0A0T6B7M3 A0A0C9RU22 A0A1B6DS29 A0A0J9RID4 B4P5G4 W8BF13 Q9W247 Q8MS45 A0A1B6LIT0 T1PD50 B3NMH7 A0A182GDL3 A0A182GC73 A0A1I8NCU7 A0A0C9Q9Q5 A0A1W4VD62 K1Q333 A0A3B0J1Z4 B3MEE5 Q28ZL4 A0A0K8UA21 J9JV32 A0A1I8NLX3 A0A1I8NLX2 A0A0A1X761 A0A034VDK8 A0A0L0BSS0 Q17NT3 B4LPJ6 B4MQG0 B4J9D3 A0A2S2R1D7 A0A0M4ET29 B3MEE4 B4KQG5 R7T326 A0A1A7WC11 E6ZIJ0 B0WIZ5 A0A1A7YDD1 A0A2M4A7B1 A0A2M4A7Q0 A0A1W4Z7H4 A0A1Q3FIQ1 A0A210QXT0 A0A182YAL1 A0A1L8E2P7 A0A182PGQ0 I3KAG2 A0A3Q3MNH3 A0A1S3K6H7 A0A3P8VR40 W5J5F0 A0A1W4Z6Z9 C3XU57 A0A1A9ZV75 A0A3Q3N2B8 I3KAG3 A0A146L8C1 A0A3P8U6D6 A0A3B3C373 A0A3P9IDB8 A0A1B0C911 A0A3Q2DXQ3 A0A1S3NHW5 A0A0K8SCA3 A0A0A9WS25 A0A3S2TZ16 A0A0B6ZGC5 U5EWT1 A0A2M4B9T0 A0A2M4B9Q2 A0A3B4XHE5 A0A3Q1BR13 A0A3B1K6I2 A0A1A8FIQ1 A0A182LW39 A0A3P9D1K7 A0A2I4C2J0 A0A3B4TD26 H2LT15 A0A1A9WNB0
A0A1Y1NFB5 A0A310SQ07 E2BGJ9 A0A232FMW6 A0A195EXZ2 E2B0R6 A0A158NLJ1 A0A1B6E0J6 A0A151IMT7 A0A195B1G5 F4WW16 A0A195E6E3 A0A0T6B7M3 A0A0C9RU22 A0A1B6DS29 A0A0J9RID4 B4P5G4 W8BF13 Q9W247 Q8MS45 A0A1B6LIT0 T1PD50 B3NMH7 A0A182GDL3 A0A182GC73 A0A1I8NCU7 A0A0C9Q9Q5 A0A1W4VD62 K1Q333 A0A3B0J1Z4 B3MEE5 Q28ZL4 A0A0K8UA21 J9JV32 A0A1I8NLX3 A0A1I8NLX2 A0A0A1X761 A0A034VDK8 A0A0L0BSS0 Q17NT3 B4LPJ6 B4MQG0 B4J9D3 A0A2S2R1D7 A0A0M4ET29 B3MEE4 B4KQG5 R7T326 A0A1A7WC11 E6ZIJ0 B0WIZ5 A0A1A7YDD1 A0A2M4A7B1 A0A2M4A7Q0 A0A1W4Z7H4 A0A1Q3FIQ1 A0A210QXT0 A0A182YAL1 A0A1L8E2P7 A0A182PGQ0 I3KAG2 A0A3Q3MNH3 A0A1S3K6H7 A0A3P8VR40 W5J5F0 A0A1W4Z6Z9 C3XU57 A0A1A9ZV75 A0A3Q3N2B8 I3KAG3 A0A146L8C1 A0A3P8U6D6 A0A3B3C373 A0A3P9IDB8 A0A1B0C911 A0A3Q2DXQ3 A0A1S3NHW5 A0A0K8SCA3 A0A0A9WS25 A0A3S2TZ16 A0A0B6ZGC5 U5EWT1 A0A2M4B9T0 A0A2M4B9Q2 A0A3B4XHE5 A0A3Q1BR13 A0A3B1K6I2 A0A1A8FIQ1 A0A182LW39 A0A3P9D1K7 A0A2I4C2J0 A0A3B4TD26 H2LT15 A0A1A9WNB0
Pubmed
19121390
24845553
30249741
28004739
20798317
28648823
+ More
21347285 21719571 22936249 17994087 17550304 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 25315136 22992520 15632085 25830018 25348373 26108605 17510324 18057021 23254933 28812685 25244985 25186727 26383154 24487278 20920257 23761445 18563158 26823975 29451363 17554307 25401762 25329095
21347285 21719571 22936249 17994087 17550304 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26483478 25315136 22992520 15632085 25830018 25348373 26108605 17510324 18057021 23254933 28812685 25244985 25186727 26383154 24487278 20920257 23761445 18563158 26823975 29451363 17554307 25401762 25329095
EMBL
BABH01021891
BABH01021892
BABH01021893
BABH01021894
KK852423
KDR24254.1
+ More
NEVH01016325 PNF25706.1 KQ982314 KYQ58101.1 QOIP01000005 RLU22243.1 GEZM01006868 GEZM01006867 JAV95610.1 KQ760243 OAD61263.1 GL448189 EFN85192.1 NNAY01000030 OXU31798.1 KQ981914 KYN33093.1 GL444666 EFN60738.1 ADTU01019476 ADTU01019477 GEDC01005842 JAS31456.1 KQ977054 KYN06086.1 KQ976662 KYM78333.1 GL888404 EGI61605.1 KQ979579 KYN20656.1 LJIG01009364 KRT83227.1 GBYB01011096 JAG80863.1 GEDC01008817 JAS28481.1 CM002911 KMY95798.1 CM000158 EDW91795.1 GAMC01018241 GAMC01018239 JAB88314.1 AE013599 BT024193 BT050450 AAF46852.2 ABC86255.1 ACJ13157.1 AY119106 AAM50966.1 GEBQ01016429 JAT23548.1 KA646604 AFP61233.1 CH954179 EDV54916.1 JXUM01056089 JXUM01056090 JXUM01056091 KQ561896 KXJ77230.1 JXUM01054671 JXUM01054672 KQ561832 KXJ77397.1 GBYB01011108 JAG80875.1 JH817484 EKC30857.1 OUUW01000001 SPP75055.1 CH902619 EDV36551.1 CM000071 EAL25599.2 GDHF01028797 JAI23517.1 ABLF02038994 GBXI01007576 JAD06716.1 GAKP01019319 JAC39633.1 JRES01001422 KNC23028.1 CH477196 EAT48391.1 CH940648 EDW61255.1 CH963849 EDW74349.1 CH916367 EDW01414.1 GGMS01014592 MBY83795.1 CP012524 ALC40600.1 EDV36550.1 KPU76291.1 CH933808 EDW10305.1 AMQN01016120 KB312557 ELT87012.1 HADW01001661 SBP03061.1 FQ310508 CBN81874.1 DS231954 EDS28856.1 HADX01006295 SBP28527.1 GGFK01003324 MBW36645.1 GGFK01003337 MBW36658.1 GFDL01007692 JAV27353.1 NEDP02001335 OWF53547.1 GFDF01001130 JAV12954.1 AERX01010097 ADMH02002183 ETN57984.1 GG666464 EEN68431.1 GDHC01015227 JAQ03402.1 AJWK01001712 GBRD01014894 JAG50932.1 GBHO01032337 GBHO01030327 JAG11267.1 JAG13277.1 CM012456 RVE58886.1 HACG01020036 CEK66901.1 GANO01001280 JAB58591.1 GGFJ01000621 MBW49762.1 GGFJ01000622 MBW49763.1 HAEB01011487 SBQ58014.1 AXCM01008426
NEVH01016325 PNF25706.1 KQ982314 KYQ58101.1 QOIP01000005 RLU22243.1 GEZM01006868 GEZM01006867 JAV95610.1 KQ760243 OAD61263.1 GL448189 EFN85192.1 NNAY01000030 OXU31798.1 KQ981914 KYN33093.1 GL444666 EFN60738.1 ADTU01019476 ADTU01019477 GEDC01005842 JAS31456.1 KQ977054 KYN06086.1 KQ976662 KYM78333.1 GL888404 EGI61605.1 KQ979579 KYN20656.1 LJIG01009364 KRT83227.1 GBYB01011096 JAG80863.1 GEDC01008817 JAS28481.1 CM002911 KMY95798.1 CM000158 EDW91795.1 GAMC01018241 GAMC01018239 JAB88314.1 AE013599 BT024193 BT050450 AAF46852.2 ABC86255.1 ACJ13157.1 AY119106 AAM50966.1 GEBQ01016429 JAT23548.1 KA646604 AFP61233.1 CH954179 EDV54916.1 JXUM01056089 JXUM01056090 JXUM01056091 KQ561896 KXJ77230.1 JXUM01054671 JXUM01054672 KQ561832 KXJ77397.1 GBYB01011108 JAG80875.1 JH817484 EKC30857.1 OUUW01000001 SPP75055.1 CH902619 EDV36551.1 CM000071 EAL25599.2 GDHF01028797 JAI23517.1 ABLF02038994 GBXI01007576 JAD06716.1 GAKP01019319 JAC39633.1 JRES01001422 KNC23028.1 CH477196 EAT48391.1 CH940648 EDW61255.1 CH963849 EDW74349.1 CH916367 EDW01414.1 GGMS01014592 MBY83795.1 CP012524 ALC40600.1 EDV36550.1 KPU76291.1 CH933808 EDW10305.1 AMQN01016120 KB312557 ELT87012.1 HADW01001661 SBP03061.1 FQ310508 CBN81874.1 DS231954 EDS28856.1 HADX01006295 SBP28527.1 GGFK01003324 MBW36645.1 GGFK01003337 MBW36658.1 GFDL01007692 JAV27353.1 NEDP02001335 OWF53547.1 GFDF01001130 JAV12954.1 AERX01010097 ADMH02002183 ETN57984.1 GG666464 EEN68431.1 GDHC01015227 JAQ03402.1 AJWK01001712 GBRD01014894 JAG50932.1 GBHO01032337 GBHO01030327 JAG11267.1 JAG13277.1 CM012456 RVE58886.1 HACG01020036 CEK66901.1 GANO01001280 JAB58591.1 GGFJ01000621 MBW49762.1 GGFJ01000622 MBW49763.1 HAEB01011487 SBQ58014.1 AXCM01008426
Proteomes
UP000005204
UP000027135
UP000235965
UP000005203
UP000075809
UP000279307
+ More
UP000008237 UP000215335 UP000078541 UP000000311 UP000005205 UP000078542 UP000078540 UP000007755 UP000078492 UP000002282 UP000000803 UP000008711 UP000069940 UP000249989 UP000095301 UP000192221 UP000005408 UP000268350 UP000007801 UP000001819 UP000007819 UP000095300 UP000037069 UP000008820 UP000008792 UP000007798 UP000001070 UP000092553 UP000009192 UP000014760 UP000002320 UP000192224 UP000242188 UP000076408 UP000075885 UP000005207 UP000261660 UP000085678 UP000265120 UP000000673 UP000001554 UP000092445 UP000261640 UP000265080 UP000261560 UP000265200 UP000092461 UP000265020 UP000087266 UP000261360 UP000257160 UP000018467 UP000075883 UP000265160 UP000192220 UP000261420 UP000001038 UP000091820
UP000008237 UP000215335 UP000078541 UP000000311 UP000005205 UP000078542 UP000078540 UP000007755 UP000078492 UP000002282 UP000000803 UP000008711 UP000069940 UP000249989 UP000095301 UP000192221 UP000005408 UP000268350 UP000007801 UP000001819 UP000007819 UP000095300 UP000037069 UP000008820 UP000008792 UP000007798 UP000001070 UP000092553 UP000009192 UP000014760 UP000002320 UP000192224 UP000242188 UP000076408 UP000075885 UP000005207 UP000261660 UP000085678 UP000265120 UP000000673 UP000001554 UP000092445 UP000261640 UP000265080 UP000261560 UP000265200 UP000092461 UP000265020 UP000087266 UP000261360 UP000257160 UP000018467 UP000075883 UP000265160 UP000192220 UP000261420 UP000001038 UP000091820
Pfam
Interpro
IPR002821
Hydantoinase_A
+ More
IPR003692 Hydantoinase_B
IPR008040 Hydant_A_N
IPR036544 QCR7_sf
IPR003197 QCR7
IPR016130 Tyr_Pase_AS
IPR017455 Znf_FYVE-rel
IPR010569 Myotubularin-like_Pase_dom
IPR013083 Znf_RING/FYVE/PHD
IPR029021 Prot-tyrosine_phosphatase-like
IPR003595 Tyr_Pase_cat
IPR011011 Znf_FYVE_PHD
IPR000306 Znf_FYVE
IPR003692 Hydantoinase_B
IPR008040 Hydant_A_N
IPR036544 QCR7_sf
IPR003197 QCR7
IPR016130 Tyr_Pase_AS
IPR017455 Znf_FYVE-rel
IPR010569 Myotubularin-like_Pase_dom
IPR013083 Znf_RING/FYVE/PHD
IPR029021 Prot-tyrosine_phosphatase-like
IPR003595 Tyr_Pase_cat
IPR011011 Znf_FYVE_PHD
IPR000306 Znf_FYVE
Gene 3D
ProteinModelPortal
H9J4K7
A0A067RWH5
A0A2J7QAU6
A0A087ZYN6
A0A151XCJ6
A0A3L8DPT6
+ More
A0A1Y1NFB5 A0A310SQ07 E2BGJ9 A0A232FMW6 A0A195EXZ2 E2B0R6 A0A158NLJ1 A0A1B6E0J6 A0A151IMT7 A0A195B1G5 F4WW16 A0A195E6E3 A0A0T6B7M3 A0A0C9RU22 A0A1B6DS29 A0A0J9RID4 B4P5G4 W8BF13 Q9W247 Q8MS45 A0A1B6LIT0 T1PD50 B3NMH7 A0A182GDL3 A0A182GC73 A0A1I8NCU7 A0A0C9Q9Q5 A0A1W4VD62 K1Q333 A0A3B0J1Z4 B3MEE5 Q28ZL4 A0A0K8UA21 J9JV32 A0A1I8NLX3 A0A1I8NLX2 A0A0A1X761 A0A034VDK8 A0A0L0BSS0 Q17NT3 B4LPJ6 B4MQG0 B4J9D3 A0A2S2R1D7 A0A0M4ET29 B3MEE4 B4KQG5 R7T326 A0A1A7WC11 E6ZIJ0 B0WIZ5 A0A1A7YDD1 A0A2M4A7B1 A0A2M4A7Q0 A0A1W4Z7H4 A0A1Q3FIQ1 A0A210QXT0 A0A182YAL1 A0A1L8E2P7 A0A182PGQ0 I3KAG2 A0A3Q3MNH3 A0A1S3K6H7 A0A3P8VR40 W5J5F0 A0A1W4Z6Z9 C3XU57 A0A1A9ZV75 A0A3Q3N2B8 I3KAG3 A0A146L8C1 A0A3P8U6D6 A0A3B3C373 A0A3P9IDB8 A0A1B0C911 A0A3Q2DXQ3 A0A1S3NHW5 A0A0K8SCA3 A0A0A9WS25 A0A3S2TZ16 A0A0B6ZGC5 U5EWT1 A0A2M4B9T0 A0A2M4B9Q2 A0A3B4XHE5 A0A3Q1BR13 A0A3B1K6I2 A0A1A8FIQ1 A0A182LW39 A0A3P9D1K7 A0A2I4C2J0 A0A3B4TD26 H2LT15 A0A1A9WNB0
A0A1Y1NFB5 A0A310SQ07 E2BGJ9 A0A232FMW6 A0A195EXZ2 E2B0R6 A0A158NLJ1 A0A1B6E0J6 A0A151IMT7 A0A195B1G5 F4WW16 A0A195E6E3 A0A0T6B7M3 A0A0C9RU22 A0A1B6DS29 A0A0J9RID4 B4P5G4 W8BF13 Q9W247 Q8MS45 A0A1B6LIT0 T1PD50 B3NMH7 A0A182GDL3 A0A182GC73 A0A1I8NCU7 A0A0C9Q9Q5 A0A1W4VD62 K1Q333 A0A3B0J1Z4 B3MEE5 Q28ZL4 A0A0K8UA21 J9JV32 A0A1I8NLX3 A0A1I8NLX2 A0A0A1X761 A0A034VDK8 A0A0L0BSS0 Q17NT3 B4LPJ6 B4MQG0 B4J9D3 A0A2S2R1D7 A0A0M4ET29 B3MEE4 B4KQG5 R7T326 A0A1A7WC11 E6ZIJ0 B0WIZ5 A0A1A7YDD1 A0A2M4A7B1 A0A2M4A7Q0 A0A1W4Z7H4 A0A1Q3FIQ1 A0A210QXT0 A0A182YAL1 A0A1L8E2P7 A0A182PGQ0 I3KAG2 A0A3Q3MNH3 A0A1S3K6H7 A0A3P8VR40 W5J5F0 A0A1W4Z6Z9 C3XU57 A0A1A9ZV75 A0A3Q3N2B8 I3KAG3 A0A146L8C1 A0A3P8U6D6 A0A3B3C373 A0A3P9IDB8 A0A1B0C911 A0A3Q2DXQ3 A0A1S3NHW5 A0A0K8SCA3 A0A0A9WS25 A0A3S2TZ16 A0A0B6ZGC5 U5EWT1 A0A2M4B9T0 A0A2M4B9Q2 A0A3B4XHE5 A0A3Q1BR13 A0A3B1K6I2 A0A1A8FIQ1 A0A182LW39 A0A3P9D1K7 A0A2I4C2J0 A0A3B4TD26 H2LT15 A0A1A9WNB0
PDB
5L9W
E-value=7.49369e-37,
Score=391
Ontologies
PATHWAY
GO
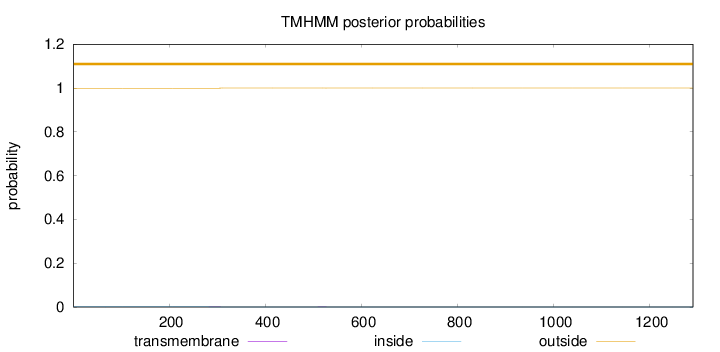
Topology
Length:
1290
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02166
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00084
outside
1 - 1290
Population Genetic Test Statistics
Pi
300.171565
Theta
176.698189
Tajima's D
2.717276
CLR
0.245717
CSRT
0.964201789910504
Interpretation
Uncertain
