Gene
KWMTBOMO11878
Annotation
ORF2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.841 Mitochondrial Reliability : 1.272
Sequence
CDS
ATGAGTGGTGTCCGGGGCGTAAGGGTCTTCCAGAATACCAGGATCGAGGAGGGCACTGTAAAAGCGGCAATAGCCGTCTTTGACGCTAATATTAGCGTTATACAGTACCCAGAACTCACCACCAATAACATCGTGGTGGTGGGGATTCGTACTGGTACCTGGGAAATCACGCTGGCATCGCTCTACTTCGAGCCGGACAAGCCTATAGAACCCTTTCTCGACCACCTAAGGAAGGTAGGAAAAGCCACCAACATCATCATCGGTGGCGATGTCAATGCCAGGAGTGTCTGGTGGGGAGGACCATCGACCAACAGCAGGGGCGAAGCGTTGCTGGGGGCCCTCGACGAGCTCGGTCTCCATGTCCTCAACCAAGGCGAAATTCCCACATTCGACGTAATCCGAGGGGAGCGACACCTGACGAGCTATGTCGATATCACGGCATGCTCACCGGATGTACTCGGCCTGGTGGACGATTGGCGAGTGCTGGAAGACCTGACGAGTTCCGACCATAATGGTATAGAGTTCAAAATTAATTTACACAAGTCCAAAGGACATACAGTACACAGAACCACACGCATATACAACACGAAAAAGGCAAATTGGGCCCAGTTTCGTGTGAAACTGGGCGAATTACTGCTGAGCACTACACTGAACAGGGACGCAATAAATAATATTAACACTATACAGGAAATTGAACAAATCACAGTAACATACACTAACACTATTACAAACGCCTGTACACACACAATACCACAAAAGAAAAATTTGGAGAAATTTTCTGTGCCATGGTGGTCGGAGGAGCTCGCCGGGTTGAAACGGCGTGTTGTCACCAGAAAGAGGAGGATCCGATGTGCCGCACTGGCCCGGAGGCCGAGGGTGGTGCAAGAGTACCTGGAGGAACGGGCAAAATACCGGGCTGCGATAAAAGAGGCCCAGATCGCAAGCTGGAAGGGGTTCTGCGAGAGGCAGGACCGCGAAGGCATGTGGGAGGGGATTTATCGGGTGATAGGCAGGGTTTCGAGACGGGAGGAAGACATACCCATGGAGCGGGAAGGGATAGTGCTGGACGCGGATAGTTCAGCCACGTACTTGGCTGAAACGTTTTATCCGGAGGACCTTACCGATTTGGACGATCCCGGGCATGGCGAGACGAGGAGGCGAGCCGCTGAACTAGATAGCGACAGTGGCGGCGCTCGAGACCCCCCGTTCACCATGCACGAGCTCGACAGATCGGTGAGGTCCTTCAACCCCAAGAAGGCTCCAGGCGCCGATGGATTGACCGCCGACATTTGTAAGGAAGCGGTCAACAGTGATCCCGAGCTGTTCCTGGCGCTCGTAAACAAGTGCCTCGAGCTGCGTTACTTCCCGAAGGTGTGGAAGGCGGCCACGGTCATCATACTCAGAAAACCTGGCAAGAACAACTACGCCACCCCCAAAGCGCACAGACCGATTGGCCTTTTGCCTGTGCTGGGGAAGGTATATGAGAAGATGATAGTGGCACGTCTAAAGTACCATATTTTGCCGAGCATGAGCGCTCACCAATATGGCTTTATGCCACAGAGGAGCACCGAAGACGCCCTATATACCCTGTTGGAACGCATTAGAGAAAAGCTTCTTGACAAGAAACTGATCCTAATGGTCTCGCTGGATATAGAGGGGGCCTTCGACAGCGCCTGGTGGCCGGCCATAAAGGTGAGGCTCGTCGAGGAGTCGTGCCCGGTTGGTATACGGGGAGTAATGAATAGTTATATGAGCGACAGGACGGTTGGAGTCCGCTACATGGGCGCCGAACATCTGAGAGCCACGGGAAAAGGATGCGTCCAGGGCTCCATAGGAGGCCCTATCCTGTGGAACCTCTTGCTCGATCCTCTGCTTAAGGGACTCAGTACCAGAGGGGACTACGTCCAGGCGTTTGCTGACGACGTTGTCCTCCTCTTCGACGGGGATACAGCGATGGACGTTCAGAGACGTGCCAATGCCGCCCTCGAGTATGTTCGGGCGTGGGGTGTCATAAACAAGCTGAAATTCGCTCCGCACAAAACCTGTGCGATGGTAATCACGCGGAAGCTGAAGCATGACACCCCGCGCCTGAGCATGGGCGGGGTAGACATTGGCATGTCTAAGGAGGTACTGTTGTTAGGGCTCACCATCGACCATAAACTTACATTTAACACCCATGTTGCCAATGTTTGCGCTAAAGCCGCAAACCTATACAAAAGGCTAACCAGGGCAGCCAAAGTTGGCTGGGGTCTGCACCCCGAAGTTATCAAGACCATATACATCGCAGTGGTTGAACCCGTGATCCTGTATGCCTCGAGCGCGTGGGCGCCGGCCGCAAACAAACTGGGAGTGCGGAAGCGCCTCAACGCGGTCCAGCGTAGCTTCGCACAGAAGCTCTGTCGAGCCTACCGAACGGTATCCCTGAACTCAGCACTGGTGCTGGCAGGGGTGCTCCCCCTTGACCTACGCGTACGCGAGATGGCCTCGCTATACGTGGCCAAGAGGGGGGCGCATCTGCCGGTGCTGGGAGACAGGGATATCGAGCGGGTGGCATCGGCACTGGAGAGGCCTCATCCAGCGGAACAAGTTGGGATGGAGCTACGAAGCCTGGTAGACGAGGAGCAATACCTCGAACATCGAGGATATGAGGTGCGTATATTTACGGACGGGAGCAAGATTGAGGGCAAGGTCGGGGCCGCGCTATCCATCTGGACTGGCGAGGCCGAAGCCAAAACCCTCAAACTTGCTCTCTCGCCCCACTGTACCGTCTACCAGGCAGAGCTCTTGGCCCTTTGTGAAGCTGCAAATGAGATCAGCCGGCGAAAAGAGAACGTTTTCGCCATCTTCAGCGACTCCATGGCGGCCCTTCAGACAGTGGCTGACCCGGATTCCCTTCACGTCCTTGCGGTCCGGATGAGGGGCGCTCTTAGTCACTGCAAAACGCAGGGCAAGAGCGTCTCCTTGTTTTGGATTAAGGCTCACGCGGGACTTGAGGGGAATGAGAGGGCCGATGCCCTGGCTAAAGCGGCCGCCCTGAGGTTGAAGAAGAAACCCAACTATGACCGGTGTCCGGTCTCATTCGCGAAGCGTGTAATCCGCATGGACACGCTCGCCGAATGGGATCGGAGGTACAGCGCCGAACAAACGGCCTCAGTCACCAAGCTGTACTTCCCAATCGCGTCCGAGGCATACAGGATAATCAGAAAGATCGAAATGACCAAGGAGGTAGCACAGGCCTTCACAGGGCATGGTGGGTTCTCCGAGTACTTGGCCAGGTTCAAGTGCAAGGAGAGCCCAGCATGCATCTGTGACCCTTACGTCGACGAGTCAGTGGCCCACGTGCTTCTGGAGTGTCCAGTCTTTGGCTCAGCACGATTCGACTTGGAGGTCGAACTGGGCCAAAGACTCCATGCGCGAGAACTCCCCAATTTAATGAAAAAAACCTGTGCGAGGAACTTGTTCCTAAAATATGCTATAAAAATTATAAAGGAGGTTAATAAAAGAAATAGAACATAA
Protein
MSGVRGVRVFQNTRIEEGTVKAAIAVFDANISVIQYPELTTNNIVVVGIRTGTWEITLASLYFEPDKPIEPFLDHLRKVGKATNIIIGGDVNARSVWWGGPSTNSRGEALLGALDELGLHVLNQGEIPTFDVIRGERHLTSYVDITACSPDVLGLVDDWRVLEDLTSSDHNGIEFKINLHKSKGHTVHRTTRIYNTKKANWAQFRVKLGELLLSTTLNRDAINNINTIQEIEQITVTYTNTITNACTHTIPQKKNLEKFSVPWWSEELAGLKRRVVTRKRRIRCAALARRPRVVQEYLEERAKYRAAIKEAQIASWKGFCERQDREGMWEGIYRVIGRVSRREEDIPMEREGIVLDADSSATYLAETFYPEDLTDLDDPGHGETRRRAAELDSDSGGARDPPFTMHELDRSVRSFNPKKAPGADGLTADICKEAVNSDPELFLALVNKCLELRYFPKVWKAATVIILRKPGKNNYATPKAHRPIGLLPVLGKVYEKMIVARLKYHILPSMSAHQYGFMPQRSTEDALYTLLERIREKLLDKKLILMVSLDIEGAFDSAWWPAIKVRLVEESCPVGIRGVMNSYMSDRTVGVRYMGAEHLRATGKGCVQGSIGGPILWNLLLDPLLKGLSTRGDYVQAFADDVVLLFDGDTAMDVQRRANAALEYVRAWGVINKLKFAPHKTCAMVITRKLKHDTPRLSMGGVDIGMSKEVLLLGLTIDHKLTFNTHVANVCAKAANLYKRLTRAAKVGWGLHPEVIKTIYIAVVEPVILYASSAWAPAANKLGVRKRLNAVQRSFAQKLCRAYRTVSLNSALVLAGVLPLDLRVREMASLYVAKRGAHLPVLGDRDIERVASALERPHPAEQVGMELRSLVDEEQYLEHRGYEVRIFTDGSKIEGKVGAALSIWTGEAEAKTLKLALSPHCTVYQAELLALCEAANEISRRKENVFAIFSDSMAALQTVADPDSLHVLAVRMRGALSHCKTQGKSVSLFWIKAHAGLEGNERADALAKAAALRLKKKPNYDRCPVSFAKRVIRMDTLAEWDRRYSAEQTASVTKLYFPIASEAYRIIRKIEMTKEVAQAFTGHGGFSEYLARFKCKESPACICDPYVDESVAHVLLECPVFGSARFDLEVELGQRLHARELPNLMKKTCARNLFLKYAIKIIKEVNKRNRT
Summary
Uniprot
Q93139
Q9BLI5
A0A224X5M4
A0A1W7R9Y4
A0A023EX80
A0A0K1IK29
+ More
A0A087TI44 A0A087TAD8 A0A3S3P0A9 A0A0K1IK14 A0A087UHT1 J9JXR0 A0A0J7KTH2 A0A1B6DRM7 A0A1B6E1N0 A0A023EYP7 A0A131XWC0 A0A1B6G0A1 J9KMZ9 U5EVA2 J9LPJ1 K7JMI9 K7JAI1 J9KPX6 A0A147BK23 A0A142LX39 J9LVC2 X1WL18 A0A224XAL8 Q5NTZ0 U5EG91 U5ERW4 A0A142LX31 A0A1I8IBT3 A0A224X696 A0A224XIB2 L7MBK1 J9LNF5 A0A142LX29 Q7M4J4 A0A224X653 J9LMF6 V9H050 J9L0V3 U5EPW8 A0A087TUG6 J9KVB7 A0A069DWT6 A0A3S2TC37 T1DCM5 J9M9N3 X1WJY4 A0A224XH41 J9K2L0 A0A142LX35 J9M2E8 J9L798 A0A142LX24 X1XHQ0 U5EW32 U5EV73 A0A142LX33 J9K5I1 J9LQV9 D7GXM9 U5ES21
A0A087TI44 A0A087TAD8 A0A3S3P0A9 A0A0K1IK14 A0A087UHT1 J9JXR0 A0A0J7KTH2 A0A1B6DRM7 A0A1B6E1N0 A0A023EYP7 A0A131XWC0 A0A1B6G0A1 J9KMZ9 U5EVA2 J9LPJ1 K7JMI9 K7JAI1 J9KPX6 A0A147BK23 A0A142LX39 J9LVC2 X1WL18 A0A224XAL8 Q5NTZ0 U5EG91 U5ERW4 A0A142LX31 A0A1I8IBT3 A0A224X696 A0A224XIB2 L7MBK1 J9LNF5 A0A142LX29 Q7M4J4 A0A224X653 J9LMF6 V9H050 J9L0V3 U5EPW8 A0A087TUG6 J9KVB7 A0A069DWT6 A0A3S2TC37 T1DCM5 J9M9N3 X1WJY4 A0A224XH41 J9K2L0 A0A142LX35 J9M2E8 J9L798 A0A142LX24 X1XHQ0 U5EW32 U5EV73 A0A142LX33 J9K5I1 J9LQV9 D7GXM9 U5ES21
Pubmed
EMBL
D38414
BAA07467.1
AB046668
BAB21511.1
GFTR01008639
JAW07787.1
+ More
GFAH01000444 JAV47945.1 GBBI01004920 JAC13792.1 KP771711 AKU04648.1 KK115320 KFM64783.1 KK114278 KFM62077.1 NCKU01007943 RWS02258.1 KP771712 AKU04650.1 KK119855 KFM76920.1 ABLF02014981 LBMM01003366 KMQ93606.1 GEDC01008950 JAS28348.1 GEDC01005462 JAS31836.1 GBBI01004973 JAC13739.1 GEFM01005915 JAP69881.1 GECZ01013919 JAS55850.1 ABLF02039177 GANO01003496 JAB56375.1 ABLF02012872 ABLF02012873 ABLF02012874 ABLF02012875 ABLF02042485 AAZX01008070 AAZX01006426 ABLF02019936 ABLF02057951 GEGO01004274 JAR91130.1 KU543680 AMS38365.1 ABLF02008516 ABLF02041501 ABLF02010037 ABLF02010039 ABLF02066755 GFTR01008452 JAW07974.1 AB182560 BAD82946.1 GANO01003539 JAB56332.1 GANO01003538 JAB56333.1 KU543676 AMS38357.1 GFTR01008431 JAW07995.1 GFTR01008645 JAW07781.1 GACK01003634 JAA61400.1 ABLF02006133 KU543675 AMS38355.1 GFTR01008469 JAW07957.1 ABLF02016436 M19755 AAC13649.1 ABLF02012458 ABLF02012459 GANO01003536 JAB56335.1 KK116791 KFM68755.1 ABLF02022279 GBGD01000371 JAC88518.1 RSAL01000474 RVE41603.1 GALA01001772 JAA93080.1 ABLF02003402 ABLF02025552 ABLF02031460 ABLF02044302 ABLF02059589 ABLF02065568 ABLF02066832 ABLF02066838 ABLF02011238 GFTR01008646 JAW07780.1 ABLF02004512 ABLF02004513 ABLF02046624 ABLF02049075 ABLF02059998 ABLF02065990 KU543678 AMS38361.1 ABLF02004774 ABLF02007045 KU543672 AMS38350.1 ABLF02010931 GANO01003141 JAB56730.1 GANO01003537 JAB56334.1 KU543677 AMS38359.1 ABLF02027069 ABLF02027071 ABLF02014982 KQ972380 EFA13507.2 GANO01003451 JAB56420.1
GFAH01000444 JAV47945.1 GBBI01004920 JAC13792.1 KP771711 AKU04648.1 KK115320 KFM64783.1 KK114278 KFM62077.1 NCKU01007943 RWS02258.1 KP771712 AKU04650.1 KK119855 KFM76920.1 ABLF02014981 LBMM01003366 KMQ93606.1 GEDC01008950 JAS28348.1 GEDC01005462 JAS31836.1 GBBI01004973 JAC13739.1 GEFM01005915 JAP69881.1 GECZ01013919 JAS55850.1 ABLF02039177 GANO01003496 JAB56375.1 ABLF02012872 ABLF02012873 ABLF02012874 ABLF02012875 ABLF02042485 AAZX01008070 AAZX01006426 ABLF02019936 ABLF02057951 GEGO01004274 JAR91130.1 KU543680 AMS38365.1 ABLF02008516 ABLF02041501 ABLF02010037 ABLF02010039 ABLF02066755 GFTR01008452 JAW07974.1 AB182560 BAD82946.1 GANO01003539 JAB56332.1 GANO01003538 JAB56333.1 KU543676 AMS38357.1 GFTR01008431 JAW07995.1 GFTR01008645 JAW07781.1 GACK01003634 JAA61400.1 ABLF02006133 KU543675 AMS38355.1 GFTR01008469 JAW07957.1 ABLF02016436 M19755 AAC13649.1 ABLF02012458 ABLF02012459 GANO01003536 JAB56335.1 KK116791 KFM68755.1 ABLF02022279 GBGD01000371 JAC88518.1 RSAL01000474 RVE41603.1 GALA01001772 JAA93080.1 ABLF02003402 ABLF02025552 ABLF02031460 ABLF02044302 ABLF02059589 ABLF02065568 ABLF02066832 ABLF02066838 ABLF02011238 GFTR01008646 JAW07780.1 ABLF02004512 ABLF02004513 ABLF02046624 ABLF02049075 ABLF02059998 ABLF02065990 KU543678 AMS38361.1 ABLF02004774 ABLF02007045 KU543672 AMS38350.1 ABLF02010931 GANO01003141 JAB56730.1 GANO01003537 JAB56334.1 KU543677 AMS38359.1 ABLF02027069 ABLF02027071 ABLF02014982 KQ972380 EFA13507.2 GANO01003451 JAB56420.1
Proteomes
Pfam
Interpro
IPR036691
Endo/exonu/phosph_ase_sf
+ More
IPR012337 RNaseH-like_sf
IPR005135 Endo/exonuclease/phosphatase
IPR000477 RT_dom
IPR036397 RNaseH_sf
IPR002156 RNaseH_domain
IPR003959 ATPase_AAA_core
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR031996 NVL2_nucleolin-bd
IPR041569 AAA_lid_3
IPR003593 AAA+_ATPase
IPR038100 NLV2_N_sf
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR007889 HTH_Psq
IPR005312 DUF1759
IPR008737 Peptidase_asp_put
IPR016187 CTDL_fold
IPR016186 C-type_lectin-like/link_sf
IPR001304 C-type_lectin-like
IPR006612 THAP_Znf
IPR012337 RNaseH-like_sf
IPR005135 Endo/exonuclease/phosphatase
IPR000477 RT_dom
IPR036397 RNaseH_sf
IPR002156 RNaseH_domain
IPR003959 ATPase_AAA_core
IPR027417 P-loop_NTPase
IPR003960 ATPase_AAA_CS
IPR031996 NVL2_nucleolin-bd
IPR041569 AAA_lid_3
IPR003593 AAA+_ATPase
IPR038100 NLV2_N_sf
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR007889 HTH_Psq
IPR005312 DUF1759
IPR008737 Peptidase_asp_put
IPR016187 CTDL_fold
IPR016186 C-type_lectin-like/link_sf
IPR001304 C-type_lectin-like
IPR006612 THAP_Znf
SUPFAM
Gene 3D
ProteinModelPortal
Q93139
Q9BLI5
A0A224X5M4
A0A1W7R9Y4
A0A023EX80
A0A0K1IK29
+ More
A0A087TI44 A0A087TAD8 A0A3S3P0A9 A0A0K1IK14 A0A087UHT1 J9JXR0 A0A0J7KTH2 A0A1B6DRM7 A0A1B6E1N0 A0A023EYP7 A0A131XWC0 A0A1B6G0A1 J9KMZ9 U5EVA2 J9LPJ1 K7JMI9 K7JAI1 J9KPX6 A0A147BK23 A0A142LX39 J9LVC2 X1WL18 A0A224XAL8 Q5NTZ0 U5EG91 U5ERW4 A0A142LX31 A0A1I8IBT3 A0A224X696 A0A224XIB2 L7MBK1 J9LNF5 A0A142LX29 Q7M4J4 A0A224X653 J9LMF6 V9H050 J9L0V3 U5EPW8 A0A087TUG6 J9KVB7 A0A069DWT6 A0A3S2TC37 T1DCM5 J9M9N3 X1WJY4 A0A224XH41 J9K2L0 A0A142LX35 J9M2E8 J9L798 A0A142LX24 X1XHQ0 U5EW32 U5EV73 A0A142LX33 J9K5I1 J9LQV9 D7GXM9 U5ES21
A0A087TI44 A0A087TAD8 A0A3S3P0A9 A0A0K1IK14 A0A087UHT1 J9JXR0 A0A0J7KTH2 A0A1B6DRM7 A0A1B6E1N0 A0A023EYP7 A0A131XWC0 A0A1B6G0A1 J9KMZ9 U5EVA2 J9LPJ1 K7JMI9 K7JAI1 J9KPX6 A0A147BK23 A0A142LX39 J9LVC2 X1WL18 A0A224XAL8 Q5NTZ0 U5EG91 U5ERW4 A0A142LX31 A0A1I8IBT3 A0A224X696 A0A224XIB2 L7MBK1 J9LNF5 A0A142LX29 Q7M4J4 A0A224X653 J9LMF6 V9H050 J9L0V3 U5EPW8 A0A087TUG6 J9KVB7 A0A069DWT6 A0A3S2TC37 T1DCM5 J9M9N3 X1WJY4 A0A224XH41 J9K2L0 A0A142LX35 J9M2E8 J9L798 A0A142LX24 X1XHQ0 U5EW32 U5EV73 A0A142LX33 J9K5I1 J9LQV9 D7GXM9 U5ES21
PDB
1WDU
E-value=2.22645e-55,
Score=550
Ontologies
GO
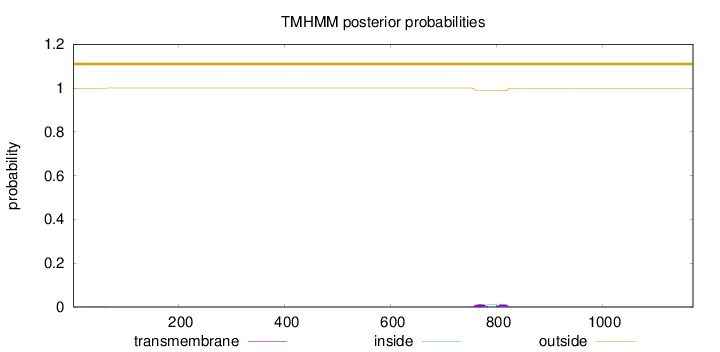
Topology
Length:
1171
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.50002
Exp number, first 60 AAs:
0.02009
Total prob of N-in:
0.00112
outside
1 - 1171
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0
CSRT
0
Interpretation
Uncertain
