Gene
KWMTBOMO11872
Pre Gene Modal
BGIBMGA004409
Annotation
PREDICTED:_dystonin-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.979 Nuclear Reliability : 1.575
Sequence
CDS
ATGCTCTCGCGATGCGACGCCATGTGGCACGATGTTTCGCAGAGTGAGAGCAAACATTTATGTTGGAACGAACGTGACGCTATACAGAAGAAGACGTTTACAAAATGGGTCAACAAACATCTCAAAAAGGTGAATCGTCACGTGAACGATCTGTTCGAGGACCTACGCGATGGACTGAACCTCATCTCGCTGCTGGAGGTGCTATCTGGCGAACATCTCCCGCGCGAGCGAGGTCGCATGCGTTTCCATATGCTGCAAAACGTCCAAATGGCCCTGGACTTCCTCCGCTACAAGAAAATCAAGCTCGTCAACATCAGAGCTGAGGATATCGTCGACGGGAACCCAAAGCTGACCCTAGGTCTGATCTGGACTATCATCCTGCACTTCCAGATATCAGACATCGTCGTTGGACAAGAACCAAACGTGACGGCCCGGGAAGCTCTCCTGAGCTGGGCCCGACGCTCCACGGCCAAGTACCCTGGGGTGAGGGTGAGCGACTTCACGTCTTCTTGGAGAGACGGGCTCGCCTTCAACGCGCTCATACACAGGAACCGCCCTGATCTGATAGACTGGCGTAATATCAGATCAAGACAAGTCCGTGAGCGCTTGGAGACAGCCTTCCATGTAGTCGAAAAGGAATATGGAGTCACCAGACTGTTGGATCCTGAAGACGTGGACACCCACGAGCCGGACGAGAAGTCCCTCATCACGTACATCTCGTCGCTGTACGAGACGTTCCCGGAGCCCCCGGCCGTGCATCCGCTGTTCGACGCGGAGTCCCAGCGCCGGGCGGCCGCTTACACCGAGCAGGCGGCCCAACATCGCGCTTGGCTGCACGAGAAATGCGCGCTAATGCAGGACCGTGCTTTCCCGTCAACTTTGATAGAAATGAAGAAGCTGCTCAGCGAGAGCACGCGGTTCCGCAACGAGGAGGTGCCGCTCAGGCAGAGAGAGAAGCAGAAGCTCGTCCACCAGTACAGGGAGTTAGAGAAATATTTCGAAACAGTGGGCGAGTGCGATATCGAGCCGACGCTGCGCCCCGAGGCGCTGGAGCAGGGCTGGGCGCGGCTGCTGATGGCGCAGCAGGAGCGGGAGCGCGACCTCACCGACGAGATTCGGCGCCTCGAGCGCCTGCAGAGACTGGCCGAGAAACTGCACAGGACAGGTGGGCGAGCAAAGGCTCAGCCAGGAGGGGTGGGATTTGCTAACAGCTACCCGAGCGCCTCCGAAGGAGACCTAACAACTCAAGAGTAG
Protein
MLSRCDAMWHDVSQSESKHLCWNERDAIQKKTFTKWVNKHLKKVNRHVNDLFEDLRDGLNLISLLEVLSGEHLPRERGRMRFHMLQNVQMALDFLRYKKIKLVNIRAEDIVDGNPKLTLGLIWTIILHFQISDIVVGQEPNVTAREALLSWARRSTAKYPGVRVSDFTSSWRDGLAFNALIHRNRPDLIDWRNIRSRQVRERLETAFHVVEKEYGVTRLLDPEDVDTHEPDEKSLITYISSLYETFPEPPAVHPLFDAESQRRAAAYTEQAAQHRAWLHEKCALMQDRAFPSTLIEMKKLLSESTRFRNEEVPLRQREKQKLVHQYRELEKYFETVGECDIEPTLRPEALEQGWARLLMAQQERERDLTDEIRRLERLQRLAEKLHRTGGRAKAQPGGVGFANSYPSASEGDLTTQE
Summary
Uniprot
A0A2A4JLN2
A0A1B6CHK9
A0A1B6DH81
A0A0J7LAV9
A0A1B6IBM2
A0A2S2QQ63
+ More
A0A1B6IXE3 A0A0C9QN10 A0A158NZ71 J9JXK1 A0A1W4VBS6 A0A1W4VBK1 A0A1W4VNK7 A0A1W4VNL7 A0A1W4VPV2 A0A1W4VPY7 A0A1W4VNJ7 A0A1W4VBV6 A0A1W4VP97 A0A1W4VPA7 A0A1W4VBL2 O77291 A0A0R1DVL9 A0A0R1DPU2 Q7KAJ2 A0A0P9C0S0 A0A0P8XRG3 A0A0P9C0N9 A0A0Q9W381 A0A0J9RCA5 A0A0J9RC92 A0A0J9RCB8 A0A0J9RDA9 A0A0P8XRF1 A0A0P8XRG7 A0A0P8XRH0 B3MHF6 A0A0P9AH49 A0A1W4VPA2 A0A0J9RDD8 A0A0J9U1A2 A0A0J9RC98 A0A0J9RC94 A0A0J9RC61 A0A0J9U195 A0A0J9U1C1 A0A0R3NT93 B5E0Y2 A0A0R3NV70 A0A0R3NN21 A0A0P8YDY3 A0A0P8XRH9 A0A0P8XRF3 A0A0J9RC91 A0A3B0JN78 A0A0R3NV84 A0A0R3NTI5 A0A0R3NMS9 A0A0R3NTT7 A0A0R3NMS3 A0A0R3NT60 A0A0R1DPS3 A0A0R1DPR4 A0A0R1DVQ3 A0A0Q5VM47 A0A0Q5VNZ6 A0A0Q5VP41 A0A0Q5W1R5 A0A0Q5VYY7 A0A0Q5VMU0 A0A0Q5VMB9 A0A0Q5VWZ7 A0A0Q5VWX9 A0A0R3NMW6 A0A0R1DPV1 A0A0R1DPR5 A0A0R1DPN8 A0A0R1DWR4 A0A0R1DPP7 A0A0Q5VME2 A0A0Q5VUT1 A0A0Q5W1M6 A0A0Q5VM70 A0A0Q5VMY8 A0A0Q5VWW2 A0A0R1DPX8 A0A0R1DPS0 A0A0R1DWS8 A0A0R1DPP6 B7YZG4 A1Z9J1 A1Z9I9 A0A0Q9XCM4 A1Z9I8 E1JH63 B7YZG2 B7YZG3 E1JH62 A0A0Q9X458 A0A0Q9X2T9
A0A1B6IXE3 A0A0C9QN10 A0A158NZ71 J9JXK1 A0A1W4VBS6 A0A1W4VBK1 A0A1W4VNK7 A0A1W4VNL7 A0A1W4VPV2 A0A1W4VPY7 A0A1W4VNJ7 A0A1W4VBV6 A0A1W4VP97 A0A1W4VPA7 A0A1W4VBL2 O77291 A0A0R1DVL9 A0A0R1DPU2 Q7KAJ2 A0A0P9C0S0 A0A0P8XRG3 A0A0P9C0N9 A0A0Q9W381 A0A0J9RCA5 A0A0J9RC92 A0A0J9RCB8 A0A0J9RDA9 A0A0P8XRF1 A0A0P8XRG7 A0A0P8XRH0 B3MHF6 A0A0P9AH49 A0A1W4VPA2 A0A0J9RDD8 A0A0J9U1A2 A0A0J9RC98 A0A0J9RC94 A0A0J9RC61 A0A0J9U195 A0A0J9U1C1 A0A0R3NT93 B5E0Y2 A0A0R3NV70 A0A0R3NN21 A0A0P8YDY3 A0A0P8XRH9 A0A0P8XRF3 A0A0J9RC91 A0A3B0JN78 A0A0R3NV84 A0A0R3NTI5 A0A0R3NMS9 A0A0R3NTT7 A0A0R3NMS3 A0A0R3NT60 A0A0R1DPS3 A0A0R1DPR4 A0A0R1DVQ3 A0A0Q5VM47 A0A0Q5VNZ6 A0A0Q5VP41 A0A0Q5W1R5 A0A0Q5VYY7 A0A0Q5VMU0 A0A0Q5VMB9 A0A0Q5VWZ7 A0A0Q5VWX9 A0A0R3NMW6 A0A0R1DPV1 A0A0R1DPR5 A0A0R1DPN8 A0A0R1DWR4 A0A0R1DPP7 A0A0Q5VME2 A0A0Q5VUT1 A0A0Q5W1M6 A0A0Q5VM70 A0A0Q5VMY8 A0A0Q5VWW2 A0A0R1DPX8 A0A0R1DPS0 A0A0R1DWS8 A0A0R1DPP6 B7YZG4 A1Z9J1 A1Z9I9 A0A0Q9XCM4 A1Z9I8 E1JH63 B7YZG2 B7YZG3 E1JH62 A0A0Q9X458 A0A0Q9X2T9
Pubmed
EMBL
NWSH01001123
PCG72484.1
GEDC01024493
JAS12805.1
GEDC01012249
JAS25049.1
+ More
LBMM01000039 KMR05210.1 GECU01032355 GECU01023404 JAS75351.1 JAS84302.1 GGMS01010467 MBY79670.1 GECU01016186 GECU01000894 JAS91520.1 JAT06813.1 GBYB01004949 JAG74716.1 ADTU01003969 ADTU01003970 ADTU01003971 ADTU01003972 ABLF02029484 AJ011924 CAA09869.1 CM000158 KRJ99257.1 KRJ99287.1 AJ011925 CAA09870.1 CH902619 KPU77163.1 KPU77142.1 KPU77127.1 CH940648 KRF79464.1 CM002911 KMY93606.1 KMY93628.1 KMY93616.1 KMY93604.1 KPU77139.1 KPU77167.1 KPU77170.1 EDV37956.1 KPU77150.1 KMY93629.1 KMY93595.1 KMY93601.1 KMY93627.1 KMY93602.1 KMY93590.1 KMY93615.1 CM000071 KRT02317.1 EDY69063.1 KRT02294.1 KRT02313.1 KPU77144.1 KPU77160.1 KPU77133.1 KMY93596.1 OUUW01000001 SPP75019.1 KRT02310.1 KRT02325.1 KRT02302.1 KRT02288.1 KRT02312.1 KRT02285.1 KRJ99274.1 KRJ99275.1 KRJ99249.1 CH954179 KQS62685.1 KQS62668.1 KQS62700.1 KQS62703.1 KQS62686.1 KQS62694.1 KQS62688.1 KQS62699.1 KQS62683.1 KRT02295.1 KRJ99260.1 KRJ99285.1 KRJ99263.1 KRJ99254.1 KRJ99269.1 KQS62704.1 KQS62677.1 KQS62671.1 KQS62701.1 KQS62673.1 KQS62667.1 KRJ99255.1 KRJ99276.1 KRJ99266.1 KRJ99295.1 KRJ99270.1 KRJ99272.1 KRJ99259.1 KRJ99267.1 AE013599 ACL83112.1 AAF58319.3 AAG22268.2 CH933808 KRG05401.1 AAF58317.1 ACZ94421.1 ACL83113.1 ACL83111.1 ACZ94420.1 CH964154 KRF99122.1 KRF99120.1
LBMM01000039 KMR05210.1 GECU01032355 GECU01023404 JAS75351.1 JAS84302.1 GGMS01010467 MBY79670.1 GECU01016186 GECU01000894 JAS91520.1 JAT06813.1 GBYB01004949 JAG74716.1 ADTU01003969 ADTU01003970 ADTU01003971 ADTU01003972 ABLF02029484 AJ011924 CAA09869.1 CM000158 KRJ99257.1 KRJ99287.1 AJ011925 CAA09870.1 CH902619 KPU77163.1 KPU77142.1 KPU77127.1 CH940648 KRF79464.1 CM002911 KMY93606.1 KMY93628.1 KMY93616.1 KMY93604.1 KPU77139.1 KPU77167.1 KPU77170.1 EDV37956.1 KPU77150.1 KMY93629.1 KMY93595.1 KMY93601.1 KMY93627.1 KMY93602.1 KMY93590.1 KMY93615.1 CM000071 KRT02317.1 EDY69063.1 KRT02294.1 KRT02313.1 KPU77144.1 KPU77160.1 KPU77133.1 KMY93596.1 OUUW01000001 SPP75019.1 KRT02310.1 KRT02325.1 KRT02302.1 KRT02288.1 KRT02312.1 KRT02285.1 KRJ99274.1 KRJ99275.1 KRJ99249.1 CH954179 KQS62685.1 KQS62668.1 KQS62700.1 KQS62703.1 KQS62686.1 KQS62694.1 KQS62688.1 KQS62699.1 KQS62683.1 KRT02295.1 KRJ99260.1 KRJ99285.1 KRJ99263.1 KRJ99254.1 KRJ99269.1 KQS62704.1 KQS62677.1 KQS62671.1 KQS62701.1 KQS62673.1 KQS62667.1 KRJ99255.1 KRJ99276.1 KRJ99266.1 KRJ99295.1 KRJ99270.1 KRJ99272.1 KRJ99259.1 KRJ99267.1 AE013599 ACL83112.1 AAF58319.3 AAG22268.2 CH933808 KRG05401.1 AAF58317.1 ACZ94421.1 ACL83113.1 ACL83111.1 ACZ94420.1 CH964154 KRF99122.1 KRF99120.1
Proteomes
Interpro
IPR002017
Spectrin_repeat
+ More
IPR001589 Actinin_actin-bd_CS
IPR001101 Plectin_repeat
IPR041615 Desmoplakin_SH3
IPR035915 Plakin_repeat_sf
IPR018159 Spectrin/alpha-actinin
IPR036872 CH_dom_sf
IPR001715 CH-domain
IPR030269 Plectin
IPR003660 HAMP_dom
IPR003108 GAR_dom
IPR036534 GAR_dom_sf
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR018247 EF_Hand_1_Ca_BS
IPR001452 SH3_domain
IPR036028 SH3-like_dom_sf
IPR001589 Actinin_actin-bd_CS
IPR001101 Plectin_repeat
IPR041615 Desmoplakin_SH3
IPR035915 Plakin_repeat_sf
IPR018159 Spectrin/alpha-actinin
IPR036872 CH_dom_sf
IPR001715 CH-domain
IPR030269 Plectin
IPR003660 HAMP_dom
IPR003108 GAR_dom
IPR036534 GAR_dom_sf
IPR011992 EF-hand-dom_pair
IPR002048 EF_hand_dom
IPR018247 EF_Hand_1_Ca_BS
IPR001452 SH3_domain
IPR036028 SH3-like_dom_sf
SUPFAM
Gene 3D
ProteinModelPortal
A0A2A4JLN2
A0A1B6CHK9
A0A1B6DH81
A0A0J7LAV9
A0A1B6IBM2
A0A2S2QQ63
+ More
A0A1B6IXE3 A0A0C9QN10 A0A158NZ71 J9JXK1 A0A1W4VBS6 A0A1W4VBK1 A0A1W4VNK7 A0A1W4VNL7 A0A1W4VPV2 A0A1W4VPY7 A0A1W4VNJ7 A0A1W4VBV6 A0A1W4VP97 A0A1W4VPA7 A0A1W4VBL2 O77291 A0A0R1DVL9 A0A0R1DPU2 Q7KAJ2 A0A0P9C0S0 A0A0P8XRG3 A0A0P9C0N9 A0A0Q9W381 A0A0J9RCA5 A0A0J9RC92 A0A0J9RCB8 A0A0J9RDA9 A0A0P8XRF1 A0A0P8XRG7 A0A0P8XRH0 B3MHF6 A0A0P9AH49 A0A1W4VPA2 A0A0J9RDD8 A0A0J9U1A2 A0A0J9RC98 A0A0J9RC94 A0A0J9RC61 A0A0J9U195 A0A0J9U1C1 A0A0R3NT93 B5E0Y2 A0A0R3NV70 A0A0R3NN21 A0A0P8YDY3 A0A0P8XRH9 A0A0P8XRF3 A0A0J9RC91 A0A3B0JN78 A0A0R3NV84 A0A0R3NTI5 A0A0R3NMS9 A0A0R3NTT7 A0A0R3NMS3 A0A0R3NT60 A0A0R1DPS3 A0A0R1DPR4 A0A0R1DVQ3 A0A0Q5VM47 A0A0Q5VNZ6 A0A0Q5VP41 A0A0Q5W1R5 A0A0Q5VYY7 A0A0Q5VMU0 A0A0Q5VMB9 A0A0Q5VWZ7 A0A0Q5VWX9 A0A0R3NMW6 A0A0R1DPV1 A0A0R1DPR5 A0A0R1DPN8 A0A0R1DWR4 A0A0R1DPP7 A0A0Q5VME2 A0A0Q5VUT1 A0A0Q5W1M6 A0A0Q5VM70 A0A0Q5VMY8 A0A0Q5VWW2 A0A0R1DPX8 A0A0R1DPS0 A0A0R1DWS8 A0A0R1DPP6 B7YZG4 A1Z9J1 A1Z9I9 A0A0Q9XCM4 A1Z9I8 E1JH63 B7YZG2 B7YZG3 E1JH62 A0A0Q9X458 A0A0Q9X2T9
A0A1B6IXE3 A0A0C9QN10 A0A158NZ71 J9JXK1 A0A1W4VBS6 A0A1W4VBK1 A0A1W4VNK7 A0A1W4VNL7 A0A1W4VPV2 A0A1W4VPY7 A0A1W4VNJ7 A0A1W4VBV6 A0A1W4VP97 A0A1W4VPA7 A0A1W4VBL2 O77291 A0A0R1DVL9 A0A0R1DPU2 Q7KAJ2 A0A0P9C0S0 A0A0P8XRG3 A0A0P9C0N9 A0A0Q9W381 A0A0J9RCA5 A0A0J9RC92 A0A0J9RCB8 A0A0J9RDA9 A0A0P8XRF1 A0A0P8XRG7 A0A0P8XRH0 B3MHF6 A0A0P9AH49 A0A1W4VPA2 A0A0J9RDD8 A0A0J9U1A2 A0A0J9RC98 A0A0J9RC94 A0A0J9RC61 A0A0J9U195 A0A0J9U1C1 A0A0R3NT93 B5E0Y2 A0A0R3NV70 A0A0R3NN21 A0A0P8YDY3 A0A0P8XRH9 A0A0P8XRF3 A0A0J9RC91 A0A3B0JN78 A0A0R3NV84 A0A0R3NTI5 A0A0R3NMS9 A0A0R3NTT7 A0A0R3NMS3 A0A0R3NT60 A0A0R1DPS3 A0A0R1DPR4 A0A0R1DVQ3 A0A0Q5VM47 A0A0Q5VNZ6 A0A0Q5VP41 A0A0Q5W1R5 A0A0Q5VYY7 A0A0Q5VMU0 A0A0Q5VMB9 A0A0Q5VWZ7 A0A0Q5VWX9 A0A0R3NMW6 A0A0R1DPV1 A0A0R1DPR5 A0A0R1DPN8 A0A0R1DWR4 A0A0R1DPP7 A0A0Q5VME2 A0A0Q5VUT1 A0A0Q5W1M6 A0A0Q5VM70 A0A0Q5VMY8 A0A0Q5VWW2 A0A0R1DPX8 A0A0R1DPS0 A0A0R1DWS8 A0A0R1DPP6 B7YZG4 A1Z9J1 A1Z9I9 A0A0Q9XCM4 A1Z9I8 E1JH63 B7YZG2 B7YZG3 E1JH62 A0A0Q9X458 A0A0Q9X2T9
PDB
4Z6G
E-value=6.56793e-95,
Score=887
Ontologies
GO
GO:0005856
GO:0003779
GO:0007010
GO:0016021
GO:0007165
GO:0005509
GO:0008017
GO:0035371
GO:0007424
GO:0035149
GO:0001578
GO:0005737
GO:0030036
GO:0016203
GO:0016319
GO:0016199
GO:0007026
GO:0035147
GO:0030175
GO:0048813
GO:0030516
GO:0045169
GO:0030426
GO:0045773
GO:0007475
GO:0000235
GO:0005874
GO:0000226
GO:0007409
GO:0007017
GO:0051491
GO:0008092
GO:0005912
GO:0016204
GO:0044295
GO:0030716
GO:0005927
GO:0007423
GO:0043622
GO:0007391
GO:0005515
GO:0006470
GO:0008138
GO:0004890
GO:0005216
GO:0043564
GO:0006418
GO:0003824
PANTHER
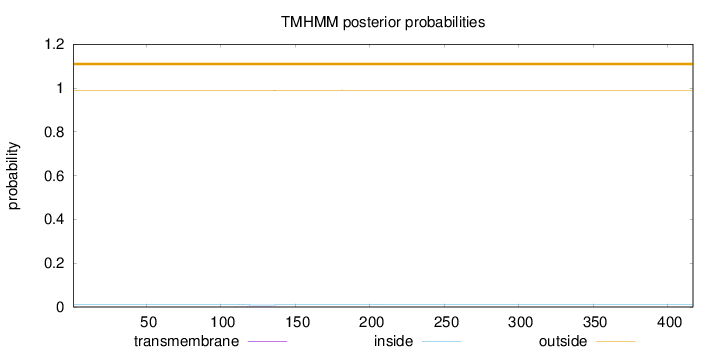
Topology
Length:
417
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.03722
Exp number, first 60 AAs:
8e-05
Total prob of N-in:
0.01148
outside
1 - 417
Population Genetic Test Statistics
Pi
182.691473
Theta
168.231388
Tajima's D
0.074609
CLR
0.253803
CSRT
0.392280385980701
Interpretation
Uncertain
