Gene
KWMTBOMO11867 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004416
Annotation
PREDICTED:_dystonin-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.298
Sequence
CDS
ATGGAGGAGGTGGGGGCGCTGGCCGCCGCGCGCCTCGCCGCGCTCGAGCACGCGCTGCCGCTCGCCGAGCACTTCGCCGACACGCACCTCGGTCTCACGTCCTGGCTGGACGACATGGAGAGGCAGATCCACATGCTGGCCATGCCCGCGCTCAGACCCGAGCAGATCGCCCAGCAGCAGGACAAGAATGAGATGCTGCAACAGTCCATAGCGGGGCACAAGCCGCTGGTGGACAAGCTGGTGAAGACGGGCGAGGCGCTGGCGCGGCTGTGCAGCGAGCTGGACGCCGCCAAGCTGCAGGACGTGCTGGAGGGAGACTGCGAGCGGTACAACGCGCTGCGGGCCGAGCTGCGGCTGCGGCAGCAGGAGCTCGAGCAGGCTCTCCAAGAATCGTCCCAGTTCTCGGACAAGCTGGAGGGGATGCTGCGGGCGCTGTCAGGCGCCGCCGACCAGCTGCAGCGCGCCGAGCCGGTCTCCGCTCACCCACCCAAGATCGAAGACCAAATCGAAGAGAACGACGCTCTGGTGGAGGACCTGGACAAGCGCAAGGAGGCGTACAACGCCGTGCAGCGCGCCGCCTCCGACGTGTTCAGCAAGGCCAACAAGAGCGACCCCGCCGTGCGCGACATCCGGAACAAACTCGACAAGTTGAACAAACTGTGGGACGAGGTGCAGCGCTCGTCGGAGAAGCGGCGGCGCGGGCTGTCGGGCGCGCTGGGCGTGGCGCGCGAGTTCTGGGCGGCGCTGCGGGCCGTGTCCAGCACGCTGGCCGAGCTCGCCGACACGCTGGCCGCGCAGCCGCCGCCCGCCGCGCACCCGCCCGCCATCCAGCAGCAGCAGCTCGCGCTGCAGGAGATCCGCCACGAGATCGACCACACCAAGCCCGAGGTGGAGAAAGTACGGAAGACCGGTTCCACTCTCATGTCTCTCTGCGGGGAACCCGACAAGCCAGAGGTCAAGAAACACATGGAGGACCTAGACAACGCATGGGACAACATCACAGCACTCTACGCCAGACGAGAGGAGAATCTCATCGATGCTATGGAAAAGGCGATGGAATTCCATGACACTTTGCAGAATCTCCAAGACTTCCTGGACACGGCGGAAGACAAGTTTTCGCGCATGGGTCCGCTCGGCTCGGACATCGACGCCGTGAAGCGTCAGATCGCTCAGCTAGCCTCCTTCAAGCAGGAGGTCGACCCTCACATGGTGAAGGTCGAAGCTTTGAACAGGCAAGCCCAGGAGCTGACAGAGAGAACGTCGTCAGAGCAAGCGGCCGCCATCAAGCAGCCCCTGACGGAGGTCAACGCTCGCTGGTCGGCCTTGCTCCGCGGCATGGTGGAGCGCCAGCAGCAACTGGAGAAGGCGCTGCTGAGACTCGGGCAGCTCCAGCACGCCCTGCAGGAGCTCCTCGCGTGGATACAGAACACCACAGACACTTTGGACACGTTGAAACCGGTGGCAGGTGACCCCCAGGTGCTGGAGGTGGAGCTGGCCAAGCTGAAGGTGCTGGTGAACGACATAGCGGCGCACCAGGCCAGCGTCGACACGCTCAACGACGCCGGCGCGCAGATCGTGCACCACGGTACCGAAGAAGCGGGCGAGACGGCCGAAAAACTGGCAACACTAAACAAAAAATGGCGCGAACTACAACAAAAAGCAAGAGACAGACAAACTGAGCTCGAGGACGCGCTCAGAGAAGCACAGAGTTTCAATGCGGAAATCGGTGACCTGCTGTCGTGGCTGTCCGAAGTGGACGGCGTCATCGCGGCCTCCAAGCCCGTCGGGGGGCTCCCGGAGACCGCCTCCGAGCAGCTGGAGAGGTTCATGGAGGTGTACAGTGAAATAGAAGAGAACCGTCCCAAAGTCGAAGCCGTGCTGCAGCAGGGACAGGAGTACATCAAGCGGCAGGAGAAACCCAACCCGACCTCGCAGCTCAACCACAGCCTCAAGACCTTGAAGTCCCGATGGGATAATGTCACTGCCAGAGCATCAGACAAGAAGATCAAGTTGGAGATAGCTCTGAAGGAGGCGACGGAGTTCCACGACGCGCTGCAGGCCTTCGTGGACTGGCTCACCGCCGCGGAGAAGACCCTCACCTCCGCCAAGCCGGTCTCCAGGGTCATGGAAACCTTGCTCACCCAAATCGAAGAGCACAAGAGCTTCCAAAAGGAGGTGTCCACGCACCGGGAGACCATGCTGCAGTTGGACAAGAAGGGAACACACCTCAAGTACTTCAGCCAGAAGCAGGACGTGATCCTCATTAAGAACTTGCTGGTCTCCGTACAGCATCGCTGGGAGAGGGTCGTCTCCAAGTCTGCCGAAAGGACCAGGGCCCTCGACCATGGCTTCAAAGAGGCTAAGGAGTTCAGTGACATGTGGAACAACCTGATGAATTGGCTGAACGAGACCGAGGAGCAGCTGGACCAACTGAACTCGGAGGCGACGGTCAACGACCCTGATAAGATCAAACAGAGATTAAACAAACACCGCGAGTTCCAAAAGGCTCTGGCTTCCAAACAGCCAGCCTACGACCTCACTATGAAGACCGGCAAGCATCTCAAGGACAAGGCTCCCAAGGGCGACGAGAACCCGCTGAAGACAATGATAACGGATATGAAGACGAAATGGACAACCGTGTGCTCTAAGGCTGTTGACAGGCAACGGAAACTGGAAGAGGCGCTGCTGTACTCGGGACAGTTCAAAGACGCCATGTCGGCGCTGCTGGACTGGCTCAAAAAGCAACAGAAGCTTCTGGAAGGAGACGCGCCTGTGCACGGCGACCTGGACACCGTGATGGCGCTGATCGAACAACATAAACAATTCGAAGAGGACCTGCATTCGCGCGAGCAGCAGATGCAGTCCGTCACCAAGACCGGCCGGGACCTGGAGGCCACCGTGCCACTGGAAGACGCCGCCAACATCCGCCAACAAGGCAGCGAACTGAAGCAACTGTGGGACAGTGTACTCATCCTCAGCGAGAGGAAAGCGCACAAGTTGGAGGGCGCACTCAAAGAGGCGGAAAAGCTGCATCGCTCTGTGAACATGCTCCTGGAGTGGCTATCGGACGCGGAGACCAAGCTTCGCTTCTCGGGCCAGCTCCCCGAAGGCGACGAGGAGACGCAGCAGCAGATACGGGACCACGAGCGCTTCGTGCGCGAACTCAACGAGAAGAAGCAGGACAAGGACGAGACCATCACGCTGGCTCACTCTATCCTGAGCAAGGCGCACACCGACGCCGTCACCGTCATCAAGCACTGGATCACCATCATACAGAGCCGCTGGGACGAGGTATGGCAATGGGCCATGCAGCGGGGCGGCAAGCTGGACGCGCACATGCAGAGCCTCAGAGACCTGGACCTCGTGCTCGAGGAGTTGCTGCAGTGGCTCACCGGACTCGAGAACAACTTGCTATCAATGGAGACTGAGCCGCTGCCTGAATCCATAGAGCTTCTCGAGGGCTTGATAGAAGACCACAAAGAACTGATGGAGCACACTCAGAAGAGACAGAACGAAGTAGACCGCGTCTGCAAAGCGTACCAGGTCAAGAGTCAAACCCAAGGAAGGGAATCTACGCCAAGGAAGGTATCAGCGAAGAGCGCCACGAAAGGAACTCCTGGCAAAGGCGCAGAACCGGAGTTCAGGTCGCCCCGCGTGAAGCAGCTGTGGGACAGGTGGCGCAACGTGTGGCTGCTCGCCTGGGAGCGCCAGCGGAGGCTGCACGAGCGCCTCGCCCATCTCAAGGAACTTCAGCGCGTCTCCAACTTCAGCTGGGACGACTGGAGGAAGAGGTTCCTCAAGTTCATGAACCACAAGAAGTCCCGCCTGACAGATCTGTTCCGTAAAATGGACAAGGACAACAACGGGCTGATACCGAGGAACGAGTTCATCGATGGCATCGTCAACACCAAATTCGACACATCCCGGCTGGAGATGGGCGCCGTGGCGGATCTGTTCGACAGGAACGGTAGCGGCCTCATCGACTGGGAGGAGTTCATTGCGGCGCTTCGCCCTGACTGGGTGGAACGCCGCGGACCCCCGACCGATGCGGACAAAATTCACGACGAGGTGAAGCGACTCGTGATGCTGTGCACGTGCCGACAGAAGTTCCGCGTGTTCCAGGTCGGAGAAGGGAAATACAGGTTCGGGGACTCCCAGAAACAGCGTCTAGTCCGTATCCTCCGCTCCACCGTCATGGTGAGGGTCGGAGGGGGCTGGGTGGCGCTCGACGAGTTCCTCGTGAAGAACGATCCCTGCAGAGCCAAGGGACGCACCAACATCGAACTGCGGGAACAGTTTATTTTGGCTGACGGTGTATCCCAGAGCATGGCAGCGTTCAGGCCGCGTACGCCTCGCTCTAACACAAATACTCCTCCATCAACCGGACCCATCACGAAGGTGAGGGAAAGAACAGCTCGATCGGTGCCTATGTCCGCTGGAGCGGGAGCGGCCGGTCGCGCTTCCAGATCTTCGTTGAGTGCAGGAACACCAGACTCGCTCAGTGATAACGAAGCGGCGAGCGGACTTGGAACAAGATACAGGAAGCCCAGCGTTCCGAGGTCCACTTTGACACCGGGAACATCTCGACCCGGTTCTCGAGCTGGATCGAGGGCAGGATCAAAACCCCCTTCGAGACATGGATCAAATTTATCCTTGGATAGTACAGACGATGCGACAACGCCGTCCCGCATTCCCATGAGGAAAGTCACGAATACGAAGACTTCGATCGCGCGTGCTGCCGCCAATGCAAGCAAGCTTGGGGTTAGCACGCCGAACGGCTCCCGTCCTCGGACGCCGACCGGTTACCTGACGCCGGCTAGTGGAAGCCCGACGCATTCCCAGCCATCTTCCATGGCATCTGAACATCCCAAGTCACTGTCCCACAATCATCCCGCATTCCAAACTCAGGGCTCGACCAAGATCCCTGTTTACGTTGGCAATCGATCTCAGAGAACTCCCTCAGTCGAGAGACGATTAAAACTGTCCAGAAAATCTAATTCCCGTGAGAATTCTCAAGAGCCCAGTCCTACCTCCCTCGATCCCTCCAGGAAGTCCGAGCAGAGGAGTCCCGAAGACGACTCATCATTGTTCAGCATATCTGGAATGACGAGTGACAATGAATACGAAAGTAATATGAGCGAGTCTAGCGGCGAAGCGTCTAAGCCGACTCAACGTGGCACCAAATGGAGTGGGTCTTTTACTTCGAACGATCTACAAACGCCTGGACGAAGTCGCATCCCAGTCCTTAAGGATCAACATACTAACAGTACTCTGAATAGACAAAGGACCCCATCGGGCAGCACGACGCCGGTGCGGTCGGGACAGCAGACCGCGGTGTCTCGTCTACTGCGCAAGCCGTCGGACGCGTCGGAGTCCGGCACGCCGGCGACGCCCGCCGCCCGCCGCGCCCCCGCCACGCCCAAAACCACAGAGAAACGCGAACCATTCCGATTGTAA
Protein
MEEVGALAAARLAALEHALPLAEHFADTHLGLTSWLDDMERQIHMLAMPALRPEQIAQQQDKNEMLQQSIAGHKPLVDKLVKTGEALARLCSELDAAKLQDVLEGDCERYNALRAELRLRQQELEQALQESSQFSDKLEGMLRALSGAADQLQRAEPVSAHPPKIEDQIEENDALVEDLDKRKEAYNAVQRAASDVFSKANKSDPAVRDIRNKLDKLNKLWDEVQRSSEKRRRGLSGALGVAREFWAALRAVSSTLAELADTLAAQPPPAAHPPAIQQQQLALQEIRHEIDHTKPEVEKVRKTGSTLMSLCGEPDKPEVKKHMEDLDNAWDNITALYARREENLIDAMEKAMEFHDTLQNLQDFLDTAEDKFSRMGPLGSDIDAVKRQIAQLASFKQEVDPHMVKVEALNRQAQELTERTSSEQAAAIKQPLTEVNARWSALLRGMVERQQQLEKALLRLGQLQHALQELLAWIQNTTDTLDTLKPVAGDPQVLEVELAKLKVLVNDIAAHQASVDTLNDAGAQIVHHGTEEAGETAEKLATLNKKWRELQQKARDRQTELEDALREAQSFNAEIGDLLSWLSEVDGVIAASKPVGGLPETASEQLERFMEVYSEIEENRPKVEAVLQQGQEYIKRQEKPNPTSQLNHSLKTLKSRWDNVTARASDKKIKLEIALKEATEFHDALQAFVDWLTAAEKTLTSAKPVSRVMETLLTQIEEHKSFQKEVSTHRETMLQLDKKGTHLKYFSQKQDVILIKNLLVSVQHRWERVVSKSAERTRALDHGFKEAKEFSDMWNNLMNWLNETEEQLDQLNSEATVNDPDKIKQRLNKHREFQKALASKQPAYDLTMKTGKHLKDKAPKGDENPLKTMITDMKTKWTTVCSKAVDRQRKLEEALLYSGQFKDAMSALLDWLKKQQKLLEGDAPVHGDLDTVMALIEQHKQFEEDLHSREQQMQSVTKTGRDLEATVPLEDAANIRQQGSELKQLWDSVLILSERKAHKLEGALKEAEKLHRSVNMLLEWLSDAETKLRFSGQLPEGDEETQQQIRDHERFVRELNEKKQDKDETITLAHSILSKAHTDAVTVIKHWITIIQSRWDEVWQWAMQRGGKLDAHMQSLRDLDLVLEELLQWLTGLENNLLSMETEPLPESIELLEGLIEDHKELMEHTQKRQNEVDRVCKAYQVKSQTQGRESTPRKVSAKSATKGTPGKGAEPEFRSPRVKQLWDRWRNVWLLAWERQRRLHERLAHLKELQRVSNFSWDDWRKRFLKFMNHKKSRLTDLFRKMDKDNNGLIPRNEFIDGIVNTKFDTSRLEMGAVADLFDRNGSGLIDWEEFIAALRPDWVERRGPPTDADKIHDEVKRLVMLCTCRQKFRVFQVGEGKYRFGDSQKQRLVRILRSTVMVRVGGGWVALDEFLVKNDPCRAKGRTNIELREQFILADGVSQSMAAFRPRTPRSNTNTPPSTGPITKVRERTARSVPMSAGAGAAGRASRSSLSAGTPDSLSDNEAASGLGTRYRKPSVPRSTLTPGTSRPGSRAGSRAGSKPPSRHGSNLSLDSTDDATTPSRIPMRKVTNTKTSIARAAANASKLGVSTPNGSRPRTPTGYLTPASGSPTHSQPSSMASEHPKSLSHNHPAFQTQGSTKIPVYVGNRSQRTPSVERRLKLSRKSNSRENSQEPSPTSLDPSRKSEQRSPEDDSSLFSISGMTSDNEYESNMSESSGEASKPTQRGTKWSGSFTSNDLQTPGRSRIPVLKDQHTNSTLNRQRTPSGSTTPVRSGQQTAVSRLLRKPSDASESGTPATPAARRAPATPKTTEKREPFRL
Summary
Uniprot
A0A3S2NXD0
H9J4H6
A0A2H1VPY4
A0A194Q5U2
A0A2J7PSP3
A0A2J7PSN8
+ More
A0A2J7PSP1 A0A2J7PSP5 A0A3L8DXH0 E2C0F1 K7JAA9 A0A195AZ78 A0A026WBN0 A0A151WQC5 A0A195F5M7 A0A151J318 A0A158NZ71 A0A1Y1KRN3 A0A1Y1KPI7 A0A1Y1KQC6 A0A087ZYV1 A0A0L7RGX0 A0A1Y1KLV0 A0A232FKV5 A0A310SII6 E2AVR2 J9JXK1 Q17GY9 A0A240PN85 A0A1S4G7Z7 A0A195D5G2 W4VR42 A0A182FBS4 Q7Q2Y5 A0A2M3ZYB6 A0A1B6C0C1 A0A084WB13 A0A0J7L314 A0A139WE75 A0A1Y1KLU3 A0A1I8MX94 A0A1Y1KU16 A0A1Y1KSF5 T1PLK8 T1PNR9 A0A1I8MXA0 A0A1I8MX89 A0A1Y1KP56 A0A2P8YWS3 A0A1I8PMG3 A0A1I8PMG2 A0A1I8MX84 A0A1I8PME4 A0A1I8PMI5 A0A1W4VNL2 B4P453 A0A1W4VPW3 A0A1W4VP93 A0A1I8PMG0 A0A0Q5VMS8 A0A1W4VPA7 A1Z9J3 A0A1W4VNI2 A0A1W4VBK1 A0A1W4VNL7 A0A1W4VP88 A0A1W4VBV6 A0A0J9RC81 A0A1W4VPY1 A0A0J9U195 A0A1I8PMG4 A0A1I8PMD4 A0A1W4VPB6 A0A1W4VPU1 A0A0J9U1A2 A0A0R1DVP9 A0A1W4VNI7 A0A1W4VNK7 A0A0P8XRH0 A0A0N8P0H8 A0A1W4VPU6 A0A0J9RCB8 A0A1W4VPB1 A0A1W4VP82 A0A1W4VPY7 A0A0J9RC99 A0A0J9RC61 A0A0R1DVT5 A0A1W4VP76 A0A0J9RC71 A0A0P8XRI4 A0A0P9C0S0 A0A0P8Y6W2 A0A0Q5W0H7 A0A1W4VNH7 B3MHF6 A0A0R1DQ15 A0A0R1DRV6 A0A0Q5VMB9 A0A0B4K869
A0A2J7PSP1 A0A2J7PSP5 A0A3L8DXH0 E2C0F1 K7JAA9 A0A195AZ78 A0A026WBN0 A0A151WQC5 A0A195F5M7 A0A151J318 A0A158NZ71 A0A1Y1KRN3 A0A1Y1KPI7 A0A1Y1KQC6 A0A087ZYV1 A0A0L7RGX0 A0A1Y1KLV0 A0A232FKV5 A0A310SII6 E2AVR2 J9JXK1 Q17GY9 A0A240PN85 A0A1S4G7Z7 A0A195D5G2 W4VR42 A0A182FBS4 Q7Q2Y5 A0A2M3ZYB6 A0A1B6C0C1 A0A084WB13 A0A0J7L314 A0A139WE75 A0A1Y1KLU3 A0A1I8MX94 A0A1Y1KU16 A0A1Y1KSF5 T1PLK8 T1PNR9 A0A1I8MXA0 A0A1I8MX89 A0A1Y1KP56 A0A2P8YWS3 A0A1I8PMG3 A0A1I8PMG2 A0A1I8MX84 A0A1I8PME4 A0A1I8PMI5 A0A1W4VNL2 B4P453 A0A1W4VPW3 A0A1W4VP93 A0A1I8PMG0 A0A0Q5VMS8 A0A1W4VPA7 A1Z9J3 A0A1W4VNI2 A0A1W4VBK1 A0A1W4VNL7 A0A1W4VP88 A0A1W4VBV6 A0A0J9RC81 A0A1W4VPY1 A0A0J9U195 A0A1I8PMG4 A0A1I8PMD4 A0A1W4VPB6 A0A1W4VPU1 A0A0J9U1A2 A0A0R1DVP9 A0A1W4VNI7 A0A1W4VNK7 A0A0P8XRH0 A0A0N8P0H8 A0A1W4VPU6 A0A0J9RCB8 A0A1W4VPB1 A0A1W4VP82 A0A1W4VPY7 A0A0J9RC99 A0A0J9RC61 A0A0R1DVT5 A0A1W4VP76 A0A0J9RC71 A0A0P8XRI4 A0A0P9C0S0 A0A0P8Y6W2 A0A0Q5W0H7 A0A1W4VNH7 B3MHF6 A0A0R1DQ15 A0A0R1DRV6 A0A0Q5VMB9 A0A0B4K869
Pubmed
EMBL
RSAL01000114
RVE46942.1
BABH01021856
ODYU01003468
SOQ42294.1
KQ459460
+ More
KPJ00724.1 NEVH01021925 PNF19352.1 PNF19348.1 PNF19350.1 PNF19349.1 QOIP01000003 RLU24639.1 GL451770 EFN78610.1 AAZX01002418 AAZX01007812 AAZX01009061 AAZX01009437 AAZX01010952 AAZX01012127 AAZX01018553 AAZX01021266 AAZX01021924 AAZX01022586 KQ976701 KYM77340.1 KK107295 EZA53051.1 KQ982843 KYQ50007.1 KQ981768 KYN35895.1 KQ980325 KYN16553.1 ADTU01003969 ADTU01003970 ADTU01003971 ADTU01003972 GEZM01079846 JAV62365.1 GEZM01079840 JAV62371.1 GEZM01079841 JAV62370.1 KQ414596 KOC70079.1 GEZM01079839 JAV62372.1 NNAY01000085 OXU31139.1 KQ759870 OAD62441.1 GL443183 EFN62471.1 ABLF02029484 CH477254 EAT45935.1 KQ976818 KYN08130.1 GANO01004316 JAB55555.1 AAAB01008966 EAA12983.4 GGFK01000174 MBW33495.1 GEDC01030331 JAS06967.1 ATLV01022301 ATLV01022302 KE525331 KFB47407.1 LBMM01001022 KMQ97051.1 KQ971354 KYB26246.1 GEZM01079842 JAV62369.1 GEZM01079844 JAV62367.1 GEZM01079845 JAV62366.1 KA649599 AFP64228.1 KA649598 AFP64227.1 GEZM01079843 JAV62368.1 PYGN01000314 PSN48675.1 CM000158 EDW90563.1 KRJ99279.1 CH954179 KQS62675.1 AE013599 AAF58320.2 CM002911 KMY93617.1 KMY93590.1 KMY93595.1 KRJ99289.1 CH902619 KPU77170.1 KPU77164.1 KMY93616.1 KMY93632.1 KMY93602.1 KRJ99281.1 KMY93603.1 KPU77169.1 KPU77163.1 KPU77138.1 KQS62674.1 EDV37956.1 KRJ99268.1 KRJ99277.1 KQS62688.1 AFH08061.1 AFH08067.1
KPJ00724.1 NEVH01021925 PNF19352.1 PNF19348.1 PNF19350.1 PNF19349.1 QOIP01000003 RLU24639.1 GL451770 EFN78610.1 AAZX01002418 AAZX01007812 AAZX01009061 AAZX01009437 AAZX01010952 AAZX01012127 AAZX01018553 AAZX01021266 AAZX01021924 AAZX01022586 KQ976701 KYM77340.1 KK107295 EZA53051.1 KQ982843 KYQ50007.1 KQ981768 KYN35895.1 KQ980325 KYN16553.1 ADTU01003969 ADTU01003970 ADTU01003971 ADTU01003972 GEZM01079846 JAV62365.1 GEZM01079840 JAV62371.1 GEZM01079841 JAV62370.1 KQ414596 KOC70079.1 GEZM01079839 JAV62372.1 NNAY01000085 OXU31139.1 KQ759870 OAD62441.1 GL443183 EFN62471.1 ABLF02029484 CH477254 EAT45935.1 KQ976818 KYN08130.1 GANO01004316 JAB55555.1 AAAB01008966 EAA12983.4 GGFK01000174 MBW33495.1 GEDC01030331 JAS06967.1 ATLV01022301 ATLV01022302 KE525331 KFB47407.1 LBMM01001022 KMQ97051.1 KQ971354 KYB26246.1 GEZM01079842 JAV62369.1 GEZM01079844 JAV62367.1 GEZM01079845 JAV62366.1 KA649599 AFP64228.1 KA649598 AFP64227.1 GEZM01079843 JAV62368.1 PYGN01000314 PSN48675.1 CM000158 EDW90563.1 KRJ99279.1 CH954179 KQS62675.1 AE013599 AAF58320.2 CM002911 KMY93617.1 KMY93590.1 KMY93595.1 KRJ99289.1 CH902619 KPU77170.1 KPU77164.1 KMY93616.1 KMY93632.1 KMY93602.1 KRJ99281.1 KMY93603.1 KPU77169.1 KPU77163.1 KPU77138.1 KQS62674.1 EDV37956.1 KRJ99268.1 KRJ99277.1 KQS62688.1 AFH08061.1 AFH08067.1
Proteomes
UP000283053
UP000005204
UP000053268
UP000235965
UP000279307
UP000008237
+ More
UP000002358 UP000078540 UP000053097 UP000075809 UP000078541 UP000078492 UP000005205 UP000005203 UP000053825 UP000215335 UP000000311 UP000007819 UP000008820 UP000075885 UP000078542 UP000069272 UP000007062 UP000030765 UP000036403 UP000007266 UP000095301 UP000245037 UP000095300 UP000192221 UP000002282 UP000008711 UP000000803 UP000007801
UP000002358 UP000078540 UP000053097 UP000075809 UP000078541 UP000078492 UP000005205 UP000005203 UP000053825 UP000215335 UP000000311 UP000007819 UP000008820 UP000075885 UP000078542 UP000069272 UP000007062 UP000030765 UP000036403 UP000007266 UP000095301 UP000245037 UP000095300 UP000192221 UP000002282 UP000008711 UP000000803 UP000007801
Pfam
Interpro
IPR002048
EF_hand_dom
+ More
IPR011992 EF-hand-dom_pair
IPR029926 Dystonin-like
IPR003108 GAR_dom
IPR002017 Spectrin_repeat
IPR036534 GAR_dom_sf
IPR018247 EF_Hand_1_Ca_BS
IPR018159 Spectrin/alpha-actinin
IPR001101 Plectin_repeat
IPR035915 Plakin_repeat_sf
IPR041615 Desmoplakin_SH3
IPR001589 Actinin_actin-bd_CS
IPR036872 CH_dom_sf
IPR001715 CH-domain
IPR001452 SH3_domain
IPR031311 CHIT_BIND_RR_consensus
IPR000618 Insect_cuticle
IPR011992 EF-hand-dom_pair
IPR029926 Dystonin-like
IPR003108 GAR_dom
IPR002017 Spectrin_repeat
IPR036534 GAR_dom_sf
IPR018247 EF_Hand_1_Ca_BS
IPR018159 Spectrin/alpha-actinin
IPR001101 Plectin_repeat
IPR035915 Plakin_repeat_sf
IPR041615 Desmoplakin_SH3
IPR001589 Actinin_actin-bd_CS
IPR036872 CH_dom_sf
IPR001715 CH-domain
IPR001452 SH3_domain
IPR031311 CHIT_BIND_RR_consensus
IPR000618 Insect_cuticle
Gene 3D
ProteinModelPortal
A0A3S2NXD0
H9J4H6
A0A2H1VPY4
A0A194Q5U2
A0A2J7PSP3
A0A2J7PSN8
+ More
A0A2J7PSP1 A0A2J7PSP5 A0A3L8DXH0 E2C0F1 K7JAA9 A0A195AZ78 A0A026WBN0 A0A151WQC5 A0A195F5M7 A0A151J318 A0A158NZ71 A0A1Y1KRN3 A0A1Y1KPI7 A0A1Y1KQC6 A0A087ZYV1 A0A0L7RGX0 A0A1Y1KLV0 A0A232FKV5 A0A310SII6 E2AVR2 J9JXK1 Q17GY9 A0A240PN85 A0A1S4G7Z7 A0A195D5G2 W4VR42 A0A182FBS4 Q7Q2Y5 A0A2M3ZYB6 A0A1B6C0C1 A0A084WB13 A0A0J7L314 A0A139WE75 A0A1Y1KLU3 A0A1I8MX94 A0A1Y1KU16 A0A1Y1KSF5 T1PLK8 T1PNR9 A0A1I8MXA0 A0A1I8MX89 A0A1Y1KP56 A0A2P8YWS3 A0A1I8PMG3 A0A1I8PMG2 A0A1I8MX84 A0A1I8PME4 A0A1I8PMI5 A0A1W4VNL2 B4P453 A0A1W4VPW3 A0A1W4VP93 A0A1I8PMG0 A0A0Q5VMS8 A0A1W4VPA7 A1Z9J3 A0A1W4VNI2 A0A1W4VBK1 A0A1W4VNL7 A0A1W4VP88 A0A1W4VBV6 A0A0J9RC81 A0A1W4VPY1 A0A0J9U195 A0A1I8PMG4 A0A1I8PMD4 A0A1W4VPB6 A0A1W4VPU1 A0A0J9U1A2 A0A0R1DVP9 A0A1W4VNI7 A0A1W4VNK7 A0A0P8XRH0 A0A0N8P0H8 A0A1W4VPU6 A0A0J9RCB8 A0A1W4VPB1 A0A1W4VP82 A0A1W4VPY7 A0A0J9RC99 A0A0J9RC61 A0A0R1DVT5 A0A1W4VP76 A0A0J9RC71 A0A0P8XRI4 A0A0P9C0S0 A0A0P8Y6W2 A0A0Q5W0H7 A0A1W4VNH7 B3MHF6 A0A0R1DQ15 A0A0R1DRV6 A0A0Q5VMB9 A0A0B4K869
A0A2J7PSP1 A0A2J7PSP5 A0A3L8DXH0 E2C0F1 K7JAA9 A0A195AZ78 A0A026WBN0 A0A151WQC5 A0A195F5M7 A0A151J318 A0A158NZ71 A0A1Y1KRN3 A0A1Y1KPI7 A0A1Y1KQC6 A0A087ZYV1 A0A0L7RGX0 A0A1Y1KLV0 A0A232FKV5 A0A310SII6 E2AVR2 J9JXK1 Q17GY9 A0A240PN85 A0A1S4G7Z7 A0A195D5G2 W4VR42 A0A182FBS4 Q7Q2Y5 A0A2M3ZYB6 A0A1B6C0C1 A0A084WB13 A0A0J7L314 A0A139WE75 A0A1Y1KLU3 A0A1I8MX94 A0A1Y1KU16 A0A1Y1KSF5 T1PLK8 T1PNR9 A0A1I8MXA0 A0A1I8MX89 A0A1Y1KP56 A0A2P8YWS3 A0A1I8PMG3 A0A1I8PMG2 A0A1I8MX84 A0A1I8PME4 A0A1I8PMI5 A0A1W4VNL2 B4P453 A0A1W4VPW3 A0A1W4VP93 A0A1I8PMG0 A0A0Q5VMS8 A0A1W4VPA7 A1Z9J3 A0A1W4VNI2 A0A1W4VBK1 A0A1W4VNL7 A0A1W4VP88 A0A1W4VBV6 A0A0J9RC81 A0A1W4VPY1 A0A0J9U195 A0A1I8PMG4 A0A1I8PMD4 A0A1W4VPB6 A0A1W4VPU1 A0A0J9U1A2 A0A0R1DVP9 A0A1W4VNI7 A0A1W4VNK7 A0A0P8XRH0 A0A0N8P0H8 A0A1W4VPU6 A0A0J9RCB8 A0A1W4VPB1 A0A1W4VP82 A0A1W4VPY7 A0A0J9RC99 A0A0J9RC61 A0A0R1DVT5 A0A1W4VP76 A0A0J9RC71 A0A0P8XRI4 A0A0P9C0S0 A0A0P8Y6W2 A0A0Q5W0H7 A0A1W4VNH7 B3MHF6 A0A0R1DQ15 A0A0R1DRV6 A0A0Q5VMB9 A0A0B4K869
PDB
5X57
E-value=1.8442e-28,
Score=320
Ontologies
GO
GO:0008017
GO:0005509
GO:0005856
GO:0003779
GO:0016021
GO:0016020
GO:0042060
GO:0005198
GO:0005737
GO:0030056
GO:0045104
GO:0042302
GO:0000235
GO:0045773
GO:0051491
GO:0044295
GO:0007391
GO:0005927
GO:0030716
GO:0035149
GO:0035371
GO:0016319
GO:0001578
GO:0007026
GO:0016199
GO:0048813
GO:0045169
GO:0030175
GO:0035147
GO:0043622
GO:0030516
GO:0007423
GO:0008092
GO:0005912
GO:0016204
GO:0016203
GO:0007409
GO:0007017
GO:0005874
GO:0030036
GO:0000226
GO:0007475
GO:0030426
GO:0007424
GO:0005515
GO:0006470
GO:0008138
GO:0004890
GO:0005216
GO:0043564
GO:0006418
GO:0003824
PANTHER
Topology
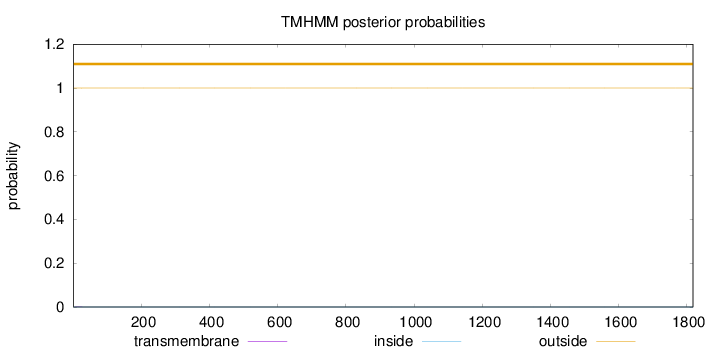
Length:
1818
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00385
Exp number, first 60 AAs:
0.00347
Total prob of N-in:
0.00019
outside
1 - 1818
Population Genetic Test Statistics
Pi
176.062483
Theta
169.920022
Tajima's D
-0.755493
CLR
0.153362
CSRT
0.184640767961602
Interpretation
Uncertain
