Pre Gene Modal
BGIBMGA004419
Annotation
proteasome_26S_non-ATPase_subunit_9_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.686
Sequence
CDS
ATGGTTAATTACAAAATAGACCCAGCTACAAGAGAATTCGTGATGAAGCTAATGGAAGAAAAAGATAGAATCGAACATTTGATTCGTGGTCATTACGCCGTATTAGCGTCCAACAATGTTGGCTTGAAGGGTTCTTTAGTTGATGAGCTAGGATACCCTCGTGATGACATAGACGTTTATGAAGTGAGACACGCGCGTCACAAGATAATTTGCCTACAAAATGATCACAAGAAAGTAATGCAACTGATCGAGAGAGGTATTGCTAAGGTTTACGAAGATTTAATTGACAGTCCAGGTATCGACAGTGAAGAGATAAACTCATGCTTAAATGGCTATCCAGTCTTTAAGAAGGATGAGACTGTCAATGATCCAACTTTTGCTACTATCAGTTTTGTGGATAAAGGTTCACCGGCTGAAGAGGCTGGTCTTCGTGCTCACGATGAACTGGTCCAATTTGGTTCAGTAAACTACAAGAACTTCAAAGATGTCTCGCAGATTATGAGAATTGTGTCACACTCTATCAATTATGGCATCACAGTTATTGTTAGGAGGGAAAATGCTGACCTAACCTTCGAGCTTGTGCCAAAACCCTGGGCTAAGCCTGGTTTACTGGGATGTCAGATACAAACAAAGAACACTTCTTAA
Protein
MVNYKIDPATREFVMKLMEEKDRIEHLIRGHYAVLASNNVGLKGSLVDELGYPRDDIDVYEVRHARHKIICLQNDHKKVMQLIERGIAKVYEDLIDSPGIDSEEINSCLNGYPVFKKDETVNDPTFATISFVDKGSPAEEAGLRAHDELVQFGSVNYKNFKDVSQIMRIVSHSINYGITVIVRRENADLTFELVPKPWAKPGLLGCQIQTKNTS
Summary
Uniprot
Q2F5N2
A0A2A4JFW9
A0A212FP20
A0A2W1BTE8
A0A2H1VNA1
A0A3S2NGG2
+ More
S4P5H4 A0A0N1IP72 E0VIY5 A0A3R7SVY6 A0A0P4W3E8 A0A1B6JR76 A0A0K8TRU9 U4UL77 J3JWE8 U5ER96 N6SYB2 A0A1Q3FIV7 A0A1Y1NHY6 A0A1B6CZX7 B0WGH0 L7M304 H2ZLM0 Q16HV7 L7M6H4 A0A1B6F5D3 A0A1E1XBK7 A0A026WUB7 A0A1S4G0P1 E2BPG9 A0A336MYW6 A0A195CEP3 A0A1Z5L485 A0A1W4WS41 A0A0P6I2H7 A0A1I8PS82 A0A023GFK8 A0A195EBE4 A0A0P5PFZ9 A0A224YG67 A0A1Q3FIG1 A0A023EK89 G3MSE1 A0A023FV21 A0A0J7L3J0 A0A0P5X857 A0A0N8D5W6 A0A182H0Q4 R7TG79 A0A232F4F5 A0A2A3EHN3 A0A088ABS3 A0A0C9RT85 A0A034W716 A0A1A8DN20 A0A131XCF3 A0A1A8GS72 A0A1A8G6K0 A0A1A8HQ76 A0A1A8ADJ6 G3NZE4 A0A310SJJ6 A0A1A8QY94 A0A1A8RYY9 A0A131YZG5 A0A3B4ANS7 A0A0F8BC40 K7J186 A0A195F959 H0XUI6 E9G414 A0A158NC84 A0A2I4BYJ6 A0A3Q1IJ41 H2XLA2 A0A3P8U8G4 A0A0Q3MMB4 A0A0N0U692 Q4RT46 A0A293N3I9 C1BW95 A0A3Q1FV71 A0A1A8CIA6 A0A1W5A6F6 A0A3Q1C3W3 A0A151WFJ0 A0A1W3JIK8 A0A1A7Z1A7 A0A1S3W227 A0A0K8VAZ0 A0A2H6NDJ4 A0A3B3B4Y5 A0A3P9HCS3 E9ILY3 W4XCS9 A0A3P9MHN0 A0A2U4CKI1 W8ASL7 A0A154P453 F7DHS1
S4P5H4 A0A0N1IP72 E0VIY5 A0A3R7SVY6 A0A0P4W3E8 A0A1B6JR76 A0A0K8TRU9 U4UL77 J3JWE8 U5ER96 N6SYB2 A0A1Q3FIV7 A0A1Y1NHY6 A0A1B6CZX7 B0WGH0 L7M304 H2ZLM0 Q16HV7 L7M6H4 A0A1B6F5D3 A0A1E1XBK7 A0A026WUB7 A0A1S4G0P1 E2BPG9 A0A336MYW6 A0A195CEP3 A0A1Z5L485 A0A1W4WS41 A0A0P6I2H7 A0A1I8PS82 A0A023GFK8 A0A195EBE4 A0A0P5PFZ9 A0A224YG67 A0A1Q3FIG1 A0A023EK89 G3MSE1 A0A023FV21 A0A0J7L3J0 A0A0P5X857 A0A0N8D5W6 A0A182H0Q4 R7TG79 A0A232F4F5 A0A2A3EHN3 A0A088ABS3 A0A0C9RT85 A0A034W716 A0A1A8DN20 A0A131XCF3 A0A1A8GS72 A0A1A8G6K0 A0A1A8HQ76 A0A1A8ADJ6 G3NZE4 A0A310SJJ6 A0A1A8QY94 A0A1A8RYY9 A0A131YZG5 A0A3B4ANS7 A0A0F8BC40 K7J186 A0A195F959 H0XUI6 E9G414 A0A158NC84 A0A2I4BYJ6 A0A3Q1IJ41 H2XLA2 A0A3P8U8G4 A0A0Q3MMB4 A0A0N0U692 Q4RT46 A0A293N3I9 C1BW95 A0A3Q1FV71 A0A1A8CIA6 A0A1W5A6F6 A0A3Q1C3W3 A0A151WFJ0 A0A1W3JIK8 A0A1A7Z1A7 A0A1S3W227 A0A0K8VAZ0 A0A2H6NDJ4 A0A3B3B4Y5 A0A3P9HCS3 E9ILY3 W4XCS9 A0A3P9MHN0 A0A2U4CKI1 W8ASL7 A0A154P453 F7DHS1
Pubmed
19121390
22118469
28756777
23622113
26354079
20566863
+ More
26369729 23537049 22516182 28004739 25576852 17510324 28503490 24508170 30249741 20798317 28528879 28797301 24945155 22216098 26483478 23254933 28648823 25348373 28049606 26830274 25463417 25835551 20075255 21292972 21347285 12481130 15114417 15496914 20433749 25069045 29451363 17554307 21282665 24495485 17495919
26369729 23537049 22516182 28004739 25576852 17510324 28503490 24508170 30249741 20798317 28528879 28797301 24945155 22216098 26483478 23254933 28648823 25348373 28049606 26830274 25463417 25835551 20075255 21292972 21347285 12481130 15114417 15496914 20433749 25069045 29451363 17554307 21282665 24495485 17495919
EMBL
BABH01021854
DQ311391
ABD36335.1
NWSH01001730
PCG70313.1
AGBW02003919
+ More
OWR55491.1 KZ149924 PZC77621.1 ODYU01003468 SOQ42293.1 RSAL01000114 RVE46941.1 GAIX01005544 JAA87016.1 KQ460547 KPJ14038.1 DS235215 EEB13341.1 QCYY01001453 ROT77962.1 GDRN01095264 GDRN01095263 JAI59569.1 GECU01005986 JAT01721.1 GDAI01000516 JAI17087.1 KB632263 ERL90916.1 BT127566 AEE62528.1 GANO01002901 JAB56970.1 APGK01052225 KB741211 ENN72784.1 GFDL01007613 JAV27432.1 GEZM01001813 JAV97512.1 GEDC01018320 JAS18978.1 DS231926 EDS26967.1 GACK01006338 JAA58696.1 CH478138 EAT33834.1 GACK01006335 JAA58699.1 GECZ01024394 JAS45375.1 GFAC01002697 JAT96491.1 KK107109 QOIP01000008 EZA59256.1 RLU19599.1 GL449633 EFN82373.1 UFQS01004095 UFQT01000960 UFQT01004095 SSX16170.1 SSX28207.1 SSX35496.1 KQ977873 KYM99170.1 GFJQ02004753 JAW02217.1 GDIQ01011529 JAN83208.1 GBBM01002562 JAC32856.1 KQ979236 KYN22159.1 GDIQ01129098 JAL22628.1 GFPF01005520 MAA16666.1 GFDL01007675 JAV27370.1 GAPW01004050 JAC09548.1 JO844792 AEO36409.1 GBBL01001981 JAC25339.1 LBMM01000808 KMQ97442.1 GDIP01142112 GDIP01076016 JAM27699.1 GDIQ01143486 GDIQ01122388 GDIP01065719 LRGB01000446 JAL08240.1 JAM37996.1 KZS19047.1 JXUM01101760 KQ564673 KXJ71919.1 AMQN01013183 KB310021 ELT92719.1 NNAY01000972 OXU25664.1 KZ288239 PBC31313.1 GBYB01011925 JAG81692.1 GAKP01008448 GAKP01008447 JAC50504.1 HAEA01006181 SBQ34661.1 GEFH01004314 JAP64267.1 HAEC01005940 SBQ74017.1 HAEB01020300 SBQ66827.1 HAED01001504 HAEE01009653 SBQ87349.1 HADY01013820 HAEJ01009229 SBP52305.1 KQ759796 OAD62831.1 HAEG01014663 SBR98193.1 HAEI01008881 HAEH01020253 SBS10504.1 GEDV01003984 JAP84573.1 KQ041059 KKF30982.1 KQ981727 KYN37140.1 AAQR03124448 AAQR03124449 GL733901 GL732531 EFX62012.1 EFX85888.1 ADTU01011739 LMAW01001359 KQK83693.1 KQ435733 KOX77271.1 CAAE01014999 CAG08436.1 GFWV01022967 MAA47694.1 BT078874 ACO13298.1 HADZ01014555 SBP78496.1 KQ983211 KYQ46601.1 HADW01000263 HADX01014062 SBP36294.1 GDHF01016614 JAI35700.1 IACI01087124 LAA30172.1 GL764129 EFZ18288.1 AAGJ04104379 GAMC01017408 JAB89147.1 KQ434809 KZC06696.1
OWR55491.1 KZ149924 PZC77621.1 ODYU01003468 SOQ42293.1 RSAL01000114 RVE46941.1 GAIX01005544 JAA87016.1 KQ460547 KPJ14038.1 DS235215 EEB13341.1 QCYY01001453 ROT77962.1 GDRN01095264 GDRN01095263 JAI59569.1 GECU01005986 JAT01721.1 GDAI01000516 JAI17087.1 KB632263 ERL90916.1 BT127566 AEE62528.1 GANO01002901 JAB56970.1 APGK01052225 KB741211 ENN72784.1 GFDL01007613 JAV27432.1 GEZM01001813 JAV97512.1 GEDC01018320 JAS18978.1 DS231926 EDS26967.1 GACK01006338 JAA58696.1 CH478138 EAT33834.1 GACK01006335 JAA58699.1 GECZ01024394 JAS45375.1 GFAC01002697 JAT96491.1 KK107109 QOIP01000008 EZA59256.1 RLU19599.1 GL449633 EFN82373.1 UFQS01004095 UFQT01000960 UFQT01004095 SSX16170.1 SSX28207.1 SSX35496.1 KQ977873 KYM99170.1 GFJQ02004753 JAW02217.1 GDIQ01011529 JAN83208.1 GBBM01002562 JAC32856.1 KQ979236 KYN22159.1 GDIQ01129098 JAL22628.1 GFPF01005520 MAA16666.1 GFDL01007675 JAV27370.1 GAPW01004050 JAC09548.1 JO844792 AEO36409.1 GBBL01001981 JAC25339.1 LBMM01000808 KMQ97442.1 GDIP01142112 GDIP01076016 JAM27699.1 GDIQ01143486 GDIQ01122388 GDIP01065719 LRGB01000446 JAL08240.1 JAM37996.1 KZS19047.1 JXUM01101760 KQ564673 KXJ71919.1 AMQN01013183 KB310021 ELT92719.1 NNAY01000972 OXU25664.1 KZ288239 PBC31313.1 GBYB01011925 JAG81692.1 GAKP01008448 GAKP01008447 JAC50504.1 HAEA01006181 SBQ34661.1 GEFH01004314 JAP64267.1 HAEC01005940 SBQ74017.1 HAEB01020300 SBQ66827.1 HAED01001504 HAEE01009653 SBQ87349.1 HADY01013820 HAEJ01009229 SBP52305.1 KQ759796 OAD62831.1 HAEG01014663 SBR98193.1 HAEI01008881 HAEH01020253 SBS10504.1 GEDV01003984 JAP84573.1 KQ041059 KKF30982.1 KQ981727 KYN37140.1 AAQR03124448 AAQR03124449 GL733901 GL732531 EFX62012.1 EFX85888.1 ADTU01011739 LMAW01001359 KQK83693.1 KQ435733 KOX77271.1 CAAE01014999 CAG08436.1 GFWV01022967 MAA47694.1 BT078874 ACO13298.1 HADZ01014555 SBP78496.1 KQ983211 KYQ46601.1 HADW01000263 HADX01014062 SBP36294.1 GDHF01016614 JAI35700.1 IACI01087124 LAA30172.1 GL764129 EFZ18288.1 AAGJ04104379 GAMC01017408 JAB89147.1 KQ434809 KZC06696.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000283053
UP000053240
UP000009046
+ More
UP000283509 UP000030742 UP000019118 UP000002320 UP000007875 UP000008820 UP000053097 UP000279307 UP000008237 UP000078542 UP000192223 UP000095300 UP000078492 UP000036403 UP000076858 UP000069940 UP000249989 UP000014760 UP000215335 UP000242457 UP000005203 UP000007635 UP000261520 UP000002358 UP000078541 UP000005225 UP000000305 UP000005205 UP000192220 UP000265040 UP000008144 UP000265080 UP000051836 UP000053105 UP000265140 UP000257200 UP000192224 UP000257160 UP000075809 UP000079721 UP000261560 UP000265200 UP000007110 UP000265180 UP000245320 UP000076502 UP000002280
UP000283509 UP000030742 UP000019118 UP000002320 UP000007875 UP000008820 UP000053097 UP000279307 UP000008237 UP000078542 UP000192223 UP000095300 UP000078492 UP000036403 UP000076858 UP000069940 UP000249989 UP000014760 UP000215335 UP000242457 UP000005203 UP000007635 UP000261520 UP000002358 UP000078541 UP000005225 UP000000305 UP000005205 UP000192220 UP000265040 UP000008144 UP000265080 UP000051836 UP000053105 UP000265140 UP000257200 UP000192224 UP000257160 UP000075809 UP000079721 UP000261560 UP000265200 UP000007110 UP000265180 UP000245320 UP000076502 UP000002280
PRIDE
SUPFAM
SSF50156
SSF50156
ProteinModelPortal
Q2F5N2
A0A2A4JFW9
A0A212FP20
A0A2W1BTE8
A0A2H1VNA1
A0A3S2NGG2
+ More
S4P5H4 A0A0N1IP72 E0VIY5 A0A3R7SVY6 A0A0P4W3E8 A0A1B6JR76 A0A0K8TRU9 U4UL77 J3JWE8 U5ER96 N6SYB2 A0A1Q3FIV7 A0A1Y1NHY6 A0A1B6CZX7 B0WGH0 L7M304 H2ZLM0 Q16HV7 L7M6H4 A0A1B6F5D3 A0A1E1XBK7 A0A026WUB7 A0A1S4G0P1 E2BPG9 A0A336MYW6 A0A195CEP3 A0A1Z5L485 A0A1W4WS41 A0A0P6I2H7 A0A1I8PS82 A0A023GFK8 A0A195EBE4 A0A0P5PFZ9 A0A224YG67 A0A1Q3FIG1 A0A023EK89 G3MSE1 A0A023FV21 A0A0J7L3J0 A0A0P5X857 A0A0N8D5W6 A0A182H0Q4 R7TG79 A0A232F4F5 A0A2A3EHN3 A0A088ABS3 A0A0C9RT85 A0A034W716 A0A1A8DN20 A0A131XCF3 A0A1A8GS72 A0A1A8G6K0 A0A1A8HQ76 A0A1A8ADJ6 G3NZE4 A0A310SJJ6 A0A1A8QY94 A0A1A8RYY9 A0A131YZG5 A0A3B4ANS7 A0A0F8BC40 K7J186 A0A195F959 H0XUI6 E9G414 A0A158NC84 A0A2I4BYJ6 A0A3Q1IJ41 H2XLA2 A0A3P8U8G4 A0A0Q3MMB4 A0A0N0U692 Q4RT46 A0A293N3I9 C1BW95 A0A3Q1FV71 A0A1A8CIA6 A0A1W5A6F6 A0A3Q1C3W3 A0A151WFJ0 A0A1W3JIK8 A0A1A7Z1A7 A0A1S3W227 A0A0K8VAZ0 A0A2H6NDJ4 A0A3B3B4Y5 A0A3P9HCS3 E9ILY3 W4XCS9 A0A3P9MHN0 A0A2U4CKI1 W8ASL7 A0A154P453 F7DHS1
S4P5H4 A0A0N1IP72 E0VIY5 A0A3R7SVY6 A0A0P4W3E8 A0A1B6JR76 A0A0K8TRU9 U4UL77 J3JWE8 U5ER96 N6SYB2 A0A1Q3FIV7 A0A1Y1NHY6 A0A1B6CZX7 B0WGH0 L7M304 H2ZLM0 Q16HV7 L7M6H4 A0A1B6F5D3 A0A1E1XBK7 A0A026WUB7 A0A1S4G0P1 E2BPG9 A0A336MYW6 A0A195CEP3 A0A1Z5L485 A0A1W4WS41 A0A0P6I2H7 A0A1I8PS82 A0A023GFK8 A0A195EBE4 A0A0P5PFZ9 A0A224YG67 A0A1Q3FIG1 A0A023EK89 G3MSE1 A0A023FV21 A0A0J7L3J0 A0A0P5X857 A0A0N8D5W6 A0A182H0Q4 R7TG79 A0A232F4F5 A0A2A3EHN3 A0A088ABS3 A0A0C9RT85 A0A034W716 A0A1A8DN20 A0A131XCF3 A0A1A8GS72 A0A1A8G6K0 A0A1A8HQ76 A0A1A8ADJ6 G3NZE4 A0A310SJJ6 A0A1A8QY94 A0A1A8RYY9 A0A131YZG5 A0A3B4ANS7 A0A0F8BC40 K7J186 A0A195F959 H0XUI6 E9G414 A0A158NC84 A0A2I4BYJ6 A0A3Q1IJ41 H2XLA2 A0A3P8U8G4 A0A0Q3MMB4 A0A0N0U692 Q4RT46 A0A293N3I9 C1BW95 A0A3Q1FV71 A0A1A8CIA6 A0A1W5A6F6 A0A3Q1C3W3 A0A151WFJ0 A0A1W3JIK8 A0A1A7Z1A7 A0A1S3W227 A0A0K8VAZ0 A0A2H6NDJ4 A0A3B3B4Y5 A0A3P9HCS3 E9ILY3 W4XCS9 A0A3P9MHN0 A0A2U4CKI1 W8ASL7 A0A154P453 F7DHS1
PDB
4O06
E-value=3.64045e-07,
Score=126
Ontologies
GO
PANTHER
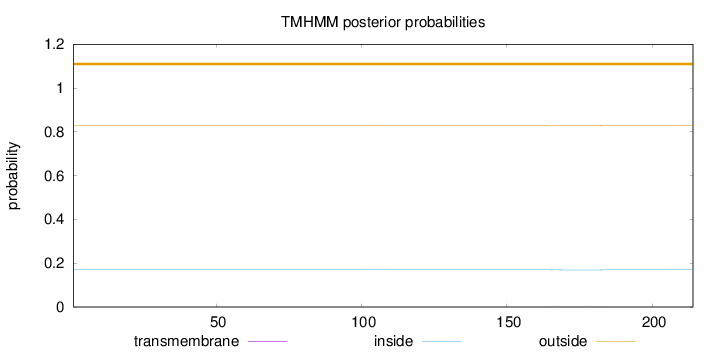
Topology
Length:
214
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.018
Exp number, first 60 AAs:
0.00201
Total prob of N-in:
0.17023
outside
1 - 214
Population Genetic Test Statistics
Pi
121.704492
Theta
153.161527
Tajima's D
-0.929556
CLR
115.367509
CSRT
0.149042547872606
Interpretation
Uncertain
