Gene
KWMTBOMO11865
Annotation
PREDICTED:_dual_specificity_protein_phosphatase_23-like_isoform_X2_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.384 Nuclear Reliability : 1.765
Sequence
CDS
ATGAGTGTTATCCGCCATACAATTTTTCTTGAGAAATTACCGCCCATCTTAGATTGTGAAAGAAAATTGAGGTGGTCCGAAATTAGAATTAAAGAGTTCGGTGCTCCATCTTTGAAGCAAATTTTAAAGTTTATTGCACTGTGTGAACGAGCGGAAATGAGAGGGGAGGTTCTAGGAGTCCATTGTCGGCATGGTCGTGGGCGCACGGGTACTATGCTGGCGTGCTATCTCGTTCACTTCAAGCGAATGGCGCCAGAACGAGCTATTCTCACGGTTAGATTGCAGAGACCAGGGTCATGTGAAACCTACGAGCAAGAAAAAGCAGTCTGTCACTACCATGACTGCGAGAGGGGAACTATAACAAAACCTGACTATCAGATGGTGGACAGCAAGCTTTATTTCGATTTTTCTATGAAATATATGTTTGACGATGACGAAACTCCAAAGCACGAGGGTAACCTGACCGACGAAACCACTGAGGAAACCAAAATAGTGTGCAAAGACCAAACCGTGAAGATTAAGCTACCTAAGAAAAAATCTAAGGCTATGGTTATCAATCATAAAATTTATTTTTAG
Protein
MSVIRHTIFLEKLPPILDCERKLRWSEIRIKEFGAPSLKQILKFIALCERAEMRGEVLGVHCRHGRGRTGTMLACYLVHFKRMAPERAILTVRLQRPGSCETYEQEKAVCHYHDCERGTITKPDYQMVDSKLYFDFSMKYMFDDDETPKHEGNLTDETTEETKIVCKDQTVKIKLPKKKSKAMVINHKIYF
Summary
Uniprot
EMBL
Proteomes
Pfam
PF00782 DSPc
Interpro
SUPFAM
SSF52799
SSF52799
Gene 3D
ProteinModelPortal
PDB
2IMG
E-value=9.63959e-22,
Score=251
Ontologies
GO
Topology
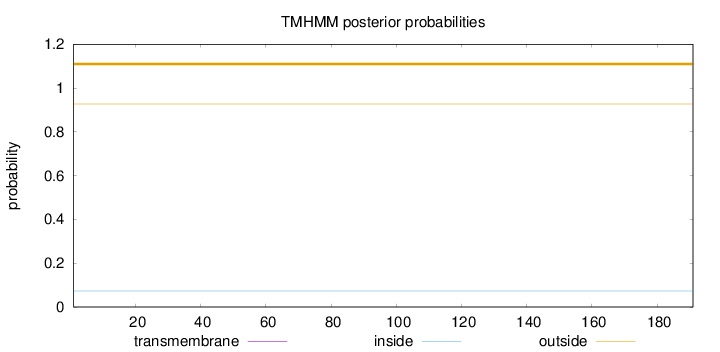
Length:
191
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00064
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07303
outside
1 - 191
Population Genetic Test Statistics
Pi
142.301849
Theta
178.460694
Tajima's D
-0.647337
CLR
0.584711
CSRT
0.203889805509725
Interpretation
Uncertain
