Gene
KWMTBOMO11864
Pre Gene Modal
BGIBMGA004441
Annotation
PREDICTED:_dual_specificity_protein_phosphatase_23-like_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 4.308
Sequence
CDS
ATGGCAGACGATGAATACCAACACATAGTGACTAGAGTAGAAGCAATAACGGAACAAGATGATAGGCATTTAGCTGGTGCAACGAAAGAAATAAGAAATGATTTAACGGTTATTATACCAGAAAATCCGTTTCCAGATATCGAAGTCGATGCGTATCCACCTCTCAAGTTTTCATGGGTCATACCTAAAAAAATTTCGGCTATGGCCTTTCCTAGGAACAAGGAGAATTTAAAATTCTTAGTAAATCAAGGCATAACTCATTTGGTGACATTAACAGCTGGAAAAAAACCTCCTGTAGACGATATTCCTAGACTGAAGTGGACAGAAGTACCTATTGAAGAGTTCGAGTTGCCGTCGGTTGAACAAATTAAGAAGTTCATGGATGTATGCAAAAGGGCTGATAAAAATGGGGAGGTATTGGGGATCCATTGCCGGCAGGGTCGGAGCAGGTCCGGCGTGATGCTGGCTTGCTACTTGGTACACTTCCACCGTTTCCTACCAGATCAAGCTGTTAATGCTATCAGAATGATAAGACCTGGGTCATGTGACTTTCCTGAGCACGAGGAAGCTGTGGGTAAATATTTCGAGTATCTCACTGAGGACAACCCTCTAAAATTCGGCGTCGGTGGAGATGTCATGGAGGAGTTCATTGACGCCGCAAAAGAGGCCACGAAGAAAGTTCTAAATTAA
Protein
MADDEYQHIVTRVEAITEQDDRHLAGATKEIRNDLTVIIPENPFPDIEVDAYPPLKFSWVIPKKISAMAFPRNKENLKFLVNQGITHLVTLTAGKKPPVDDIPRLKWTEVPIEEFELPSVEQIKKFMDVCKRADKNGEVLGIHCRQGRSRSGVMLACYLVHFHRFLPDQAVNAIRMIRPGSCDFPEHEEAVGKYFEYLTEDNPLKFGVGGDVMEEFIDAAKEATKKVLN
Summary
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF00782 DSPc
Interpro
SUPFAM
SSF52799
SSF52799
Gene 3D
ProteinModelPortal
PDB
2IMG
E-value=4.59528e-25,
Score=281
Ontologies
GO
Topology
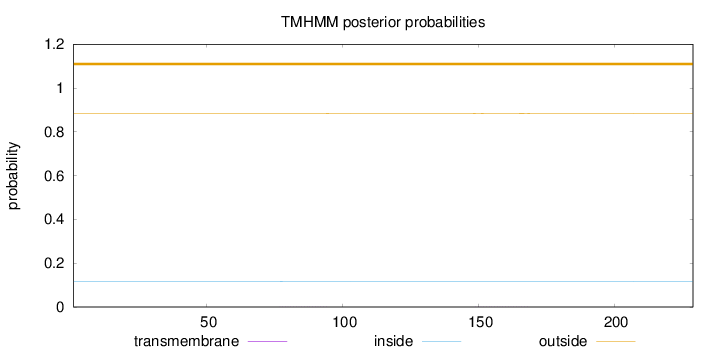
Length:
229
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02017
Exp number, first 60 AAs:
4e-05
Total prob of N-in:
0.11579
outside
1 - 229
Population Genetic Test Statistics
Pi
228.0113
Theta
191.991288
Tajima's D
0.406417
CLR
0.02245
CSRT
0.491425428728564
Interpretation
Possibly Positive selection
