Gene
KWMTBOMO11863
Pre Gene Modal
BGIBMGA004440
Annotation
PREDICTED:_partitioning_defective_protein_6_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.648 Nuclear Reliability : 2.184
Sequence
CDS
ATGTCTAAAAATAAGATAGCAGCTCGGATTGACAGTCAAACTGTCGAGGTGAAGACGAAGTTCGATGCAGAGTTCCGTCGTTTCTCGCTGAATCGTACCGAGAAACTGAAATATGACGATTTCAAAGCGCTAATCGAGAAACTGCATCGGTTACACGACGTCGCCTTCCTGATATCATACACCGATCCTAGGGACGGCGATTTACTACCCATCAACAATGATGACAACTTTGCGAGAGCATTGCTGACAGCCAGACCACTGCTCAGGGTCATAATACAAAGGAAAGGCGACAGCTTAGAAGAACTAAACGGTTATGGGACAATGCGACCGCGGAACATAATATCGTCGATCTTAAGCGGTACGCCGGCGAAGAACAAAGGACTCGCCATATCAAACCCACATGATTTTAGAATGGTGTCGGCTATAATCGACGTGGACATCGTCCCGGAGACCTGCAGGAGGGTGAAGCTGCTGAAACACGGCTCCGACAGACCATTAGGCTTCTTCATTAGGGACGGTACATCAGTACGAGTCACACCGAACGGCGTGGAGCGTTCGCCCGGTATATTTATATCGCGGCTAGTGCCAGGAGGTCTGGCGGAGTCCACGGGCCTCCTGGGGGTCAACGATGAAGTGTTGGAGGTCAACGGTATTGATGTCTCCAACAAAACACTTGATCAGGTAACGGACATGATGGTAGCCAATTCTTCGAACCTGATTATAACAGTACGTCCCGCCAATCAACGCGCCGCGCCGCACGACGCTGCGGCACACGACGCCACGCCGCACGACGCCGCGGCACACGACGCGCACACGCCCCCCGCCGACCACGACGACGACAGGTTCGATCAAGACGAGCAAGACGAGATCGTCGATCTGACCGCCGTGACGCTGGAGGAGTACCCGGAGGGCGGCGCGCGCGGCCCGGCCCGCGACGACGGACACGTGCTGCACCTCTAG
Protein
MSKNKIAARIDSQTVEVKTKFDAEFRRFSLNRTEKLKYDDFKALIEKLHRLHDVAFLISYTDPRDGDLLPINNDDNFARALLTARPLLRVIIQRKGDSLEELNGYGTMRPRNIISSILSGTPAKNKGLAISNPHDFRMVSAIIDVDIVPETCRRVKLLKHGSDRPLGFFIRDGTSVRVTPNGVERSPGIFISRLVPGGLAESTGLLGVNDEVLEVNGIDVSNKTLDQVTDMMVANSSNLIITVRPANQRAAPHDAAAHDATPHDAAAHDAHTPPADHDDDRFDQDEQDEIVDLTAVTLEEYPEGGARGPARDDGHVLHL
Summary
Similarity
Belongs to the CDP-alcohol phosphatidyltransferase class-I family.
Uniprot
A0A2W1BVG6
A0A2A4JER0
A0A3S2NRV2
A0A0N0PCK6
A0A194Q6V1
A0A2H1VN75
+ More
A0A1W4WTC1 N6T1I2 A0A1Q3G0V9 Q176B0 A0A067R9K8 A0A1B6DMV8 A0A0K8TKQ3 A0A1Y1LHI3 A0A182GHT9 A0A1B6ERF9 U5ERQ4 A0A084WAJ5 A0A182M440 A0A182YFA0 A0A182RIU8 A0A182VT53 A0A182QPF2 A0A182NDD4 A0A2M3Z781 A0A2M4AJI2 W5JSW6 A0A182FTG4 A0A182VJK6 A0A182WUX1 Q7Q8X8 A0A182HYG5 A0A2J7PSG8 A0A182JQQ6 A0A182PUG8 A0A0A9YY36 A0A1L8DMQ2 A0A139WF22 A0A0T6B8A5 V5GR04 J9KAC2 A0A2S2PNE2 A0A2H8TUE7 A0A336K1L5 A0A023F4U2 A0A069DSM6 A0A0V0G9K6 T1IAA9 A0A0P4VHM2 A0A182LPX1 A0A310SIK8 A0A182TK69 A0A0M8ZRU2 A0A1J1HI01 A0A224XS57 A0A0L7RFT8 B4H9W1 Q29JD9 A0A1W4UVQ6 B3MQV2 B4R6G0 B3NWX0 A0A3B0J919 O97111 B4Q2J3 A0A1E1WPC7 A0A0A1XDA8 A0A0K8W5G0 A0A034WFY7 B4JKB8 A0A0M3QZG7 A0A0N0BDA9 W8AT66 A0A182IQJ6 A0A088A2G7 A0A2A3EB00 B4MA03 B4L7V3 E0W0G3 A0A2J7PSG9 A0A232F0U0 A0A226EX36 A0A1D2MQQ1 B4N209 K7J0F4 E2C4Z1 A0A0R3P3L1 A0A2P6L1L1 E2AIN3 A0A154NWG6 A0A151XI98 A0A151J8G3 E9FTS2 A0A158NF92 A0A151JZY1 F4WN43 A0A151I5Z4 A0A164KBQ7 A0A2H5BF90
A0A1W4WTC1 N6T1I2 A0A1Q3G0V9 Q176B0 A0A067R9K8 A0A1B6DMV8 A0A0K8TKQ3 A0A1Y1LHI3 A0A182GHT9 A0A1B6ERF9 U5ERQ4 A0A084WAJ5 A0A182M440 A0A182YFA0 A0A182RIU8 A0A182VT53 A0A182QPF2 A0A182NDD4 A0A2M3Z781 A0A2M4AJI2 W5JSW6 A0A182FTG4 A0A182VJK6 A0A182WUX1 Q7Q8X8 A0A182HYG5 A0A2J7PSG8 A0A182JQQ6 A0A182PUG8 A0A0A9YY36 A0A1L8DMQ2 A0A139WF22 A0A0T6B8A5 V5GR04 J9KAC2 A0A2S2PNE2 A0A2H8TUE7 A0A336K1L5 A0A023F4U2 A0A069DSM6 A0A0V0G9K6 T1IAA9 A0A0P4VHM2 A0A182LPX1 A0A310SIK8 A0A182TK69 A0A0M8ZRU2 A0A1J1HI01 A0A224XS57 A0A0L7RFT8 B4H9W1 Q29JD9 A0A1W4UVQ6 B3MQV2 B4R6G0 B3NWX0 A0A3B0J919 O97111 B4Q2J3 A0A1E1WPC7 A0A0A1XDA8 A0A0K8W5G0 A0A034WFY7 B4JKB8 A0A0M3QZG7 A0A0N0BDA9 W8AT66 A0A182IQJ6 A0A088A2G7 A0A2A3EB00 B4MA03 B4L7V3 E0W0G3 A0A2J7PSG9 A0A232F0U0 A0A226EX36 A0A1D2MQQ1 B4N209 K7J0F4 E2C4Z1 A0A0R3P3L1 A0A2P6L1L1 E2AIN3 A0A154NWG6 A0A151XI98 A0A151J8G3 E9FTS2 A0A158NF92 A0A151JZY1 F4WN43 A0A151I5Z4 A0A164KBQ7 A0A2H5BF90
Pubmed
28756777
26354079
23537049
17510324
24845553
26369729
+ More
28004739 26483478 24438588 25244985 20920257 23761445 12364791 14747013 17210077 25401762 26823975 18362917 19820115 25474469 26334808 27129103 20966253 17994087 15632085 9834192 10731132 12537568 12537572 12537573 12537574 15023337 15475968 16110336 17569856 17569867 22078569 26109357 26109356 26894406 17550304 25830018 25348373 24495485 20566863 28648823 27289101 20075255 20798317 21292972 21347285 21719571 29228898
28004739 26483478 24438588 25244985 20920257 23761445 12364791 14747013 17210077 25401762 26823975 18362917 19820115 25474469 26334808 27129103 20966253 17994087 15632085 9834192 10731132 12537568 12537572 12537573 12537574 15023337 15475968 16110336 17569856 17569867 22078569 26109357 26109356 26894406 17550304 25830018 25348373 24495485 20566863 28648823 27289101 20075255 20798317 21292972 21347285 21719571 29228898
EMBL
KZ149924
PZC77624.1
NWSH01001730
PCG70309.1
RSAL01000114
RVE46939.1
+ More
KQ460547 KPJ14041.1 KQ459460 KPJ00730.1 ODYU01003468 SOQ42289.1 APGK01055954 KB741271 KB630998 KB630999 KB632114 ENN71383.1 ERL83988.1 ERL83991.1 ERL88919.1 GFDL01001606 JAV33439.1 CH477391 EAT41948.1 KK852611 KDR20262.1 GEDC01030422 GEDC01026602 GEDC01015984 GEDC01010293 JAS06876.1 JAS10696.1 JAS21314.1 JAS27005.1 GDAI01002704 JAI14899.1 GEZM01055144 JAV73112.1 JXUM01064747 JXUM01064748 KQ562313 KXJ76166.1 GECZ01029311 GECZ01023706 JAS40458.1 JAS46063.1 GANO01003620 JAB56251.1 ATLV01022200 KE525330 KFB47239.1 AXCM01000018 AXCN02001767 GGFM01003613 MBW24364.1 GGFK01007467 MBW40788.1 ADMH02000540 ETN66015.1 AAAB01008933 EAA09930.4 APCN01005907 NEVH01021928 PNF19287.1 GBHO01023939 GBHO01023938 GBHO01023937 GBHO01023935 GBHO01007596 GDHC01018450 GDHC01005022 JAG19665.1 JAG19666.1 JAG19667.1 JAG19669.1 JAG36008.1 JAQ00179.1 JAQ13607.1 GFDF01006429 JAV07655.1 KQ971354 KYB26461.1 LJIG01009185 KRT83586.1 GALX01005773 JAB62693.1 ABLF02039933 ABLF02039935 GGMR01018314 MBY30933.1 GFXV01004993 MBW16798.1 UFQS01000027 UFQT01000027 SSW97702.1 SSX18088.1 GBBI01002713 JAC15999.1 GBGD01002153 JAC86736.1 GECL01001600 JAP04524.1 ACPB03005084 GDKW01002550 JAI54045.1 KQ759843 OAD62608.1 KQ435881 KOX69840.1 CVRI01000004 CRK87523.1 GFTR01005064 JAW11362.1 KQ414601 KOC69705.1 CH479232 EDW36618.1 CH379063 EAL32362.1 CH902622 EDV34157.1 CM000366 EDX18210.1 CH954180 EDV46517.1 OUUW01000003 SPP78697.1 AF070969 AE014298 AY061369 AAD15927.1 AAF48757.1 AAL28917.1 AAN09592.1 CM000162 EDX02634.1 GDQN01002212 JAT88842.1 GBXI01005366 JAD08926.1 GDHF01006184 JAI46130.1 GAKP01005343 JAC53609.1 CH916370 EDW00021.1 CP012528 ALC49393.1 KOX69841.1 GAMC01017158 JAB89397.1 KZ288311 PBC28472.1 CH940655 EDW66062.1 CH933814 EDW05528.2 DS235858 EEB19119.1 PNF19288.1 NNAY01001423 OXU24028.1 LNIX01000001 OXA62109.1 LJIJ01000677 ODM95407.1 CH963925 EDW78398.2 GL452712 EFN76972.1 KRT05936.1 MWRG01002592 PRD32451.1 GL439840 EFN66722.1 KQ434772 KZC03892.1 KQ982080 KYQ60132.1 KQ979533 KYN21109.1 GL732524 EFX89593.1 ADTU01014064 ADTU01014065 ADTU01014066 KQ981319 KYN42718.1 GL888234 EGI64385.1 KQ976404 KYM92074.1 LRGB01003341 KZS03128.1 MG197656 AUG84406.1
KQ460547 KPJ14041.1 KQ459460 KPJ00730.1 ODYU01003468 SOQ42289.1 APGK01055954 KB741271 KB630998 KB630999 KB632114 ENN71383.1 ERL83988.1 ERL83991.1 ERL88919.1 GFDL01001606 JAV33439.1 CH477391 EAT41948.1 KK852611 KDR20262.1 GEDC01030422 GEDC01026602 GEDC01015984 GEDC01010293 JAS06876.1 JAS10696.1 JAS21314.1 JAS27005.1 GDAI01002704 JAI14899.1 GEZM01055144 JAV73112.1 JXUM01064747 JXUM01064748 KQ562313 KXJ76166.1 GECZ01029311 GECZ01023706 JAS40458.1 JAS46063.1 GANO01003620 JAB56251.1 ATLV01022200 KE525330 KFB47239.1 AXCM01000018 AXCN02001767 GGFM01003613 MBW24364.1 GGFK01007467 MBW40788.1 ADMH02000540 ETN66015.1 AAAB01008933 EAA09930.4 APCN01005907 NEVH01021928 PNF19287.1 GBHO01023939 GBHO01023938 GBHO01023937 GBHO01023935 GBHO01007596 GDHC01018450 GDHC01005022 JAG19665.1 JAG19666.1 JAG19667.1 JAG19669.1 JAG36008.1 JAQ00179.1 JAQ13607.1 GFDF01006429 JAV07655.1 KQ971354 KYB26461.1 LJIG01009185 KRT83586.1 GALX01005773 JAB62693.1 ABLF02039933 ABLF02039935 GGMR01018314 MBY30933.1 GFXV01004993 MBW16798.1 UFQS01000027 UFQT01000027 SSW97702.1 SSX18088.1 GBBI01002713 JAC15999.1 GBGD01002153 JAC86736.1 GECL01001600 JAP04524.1 ACPB03005084 GDKW01002550 JAI54045.1 KQ759843 OAD62608.1 KQ435881 KOX69840.1 CVRI01000004 CRK87523.1 GFTR01005064 JAW11362.1 KQ414601 KOC69705.1 CH479232 EDW36618.1 CH379063 EAL32362.1 CH902622 EDV34157.1 CM000366 EDX18210.1 CH954180 EDV46517.1 OUUW01000003 SPP78697.1 AF070969 AE014298 AY061369 AAD15927.1 AAF48757.1 AAL28917.1 AAN09592.1 CM000162 EDX02634.1 GDQN01002212 JAT88842.1 GBXI01005366 JAD08926.1 GDHF01006184 JAI46130.1 GAKP01005343 JAC53609.1 CH916370 EDW00021.1 CP012528 ALC49393.1 KOX69841.1 GAMC01017158 JAB89397.1 KZ288311 PBC28472.1 CH940655 EDW66062.1 CH933814 EDW05528.2 DS235858 EEB19119.1 PNF19288.1 NNAY01001423 OXU24028.1 LNIX01000001 OXA62109.1 LJIJ01000677 ODM95407.1 CH963925 EDW78398.2 GL452712 EFN76972.1 KRT05936.1 MWRG01002592 PRD32451.1 GL439840 EFN66722.1 KQ434772 KZC03892.1 KQ982080 KYQ60132.1 KQ979533 KYN21109.1 GL732524 EFX89593.1 ADTU01014064 ADTU01014065 ADTU01014066 KQ981319 KYN42718.1 GL888234 EGI64385.1 KQ976404 KYM92074.1 LRGB01003341 KZS03128.1 MG197656 AUG84406.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000053268
UP000192223
UP000019118
+ More
UP000030742 UP000008820 UP000027135 UP000069940 UP000249989 UP000030765 UP000075883 UP000076408 UP000075900 UP000075920 UP000075886 UP000075884 UP000000673 UP000069272 UP000075903 UP000076407 UP000007062 UP000075840 UP000235965 UP000075881 UP000075885 UP000007266 UP000007819 UP000015103 UP000075882 UP000075902 UP000053105 UP000183832 UP000053825 UP000008744 UP000001819 UP000192221 UP000007801 UP000000304 UP000008711 UP000268350 UP000000803 UP000002282 UP000001070 UP000092553 UP000075880 UP000005203 UP000242457 UP000008792 UP000009192 UP000009046 UP000215335 UP000198287 UP000094527 UP000007798 UP000002358 UP000008237 UP000000311 UP000076502 UP000075809 UP000078492 UP000000305 UP000005205 UP000078541 UP000007755 UP000078540 UP000076858
UP000030742 UP000008820 UP000027135 UP000069940 UP000249989 UP000030765 UP000075883 UP000076408 UP000075900 UP000075920 UP000075886 UP000075884 UP000000673 UP000069272 UP000075903 UP000076407 UP000007062 UP000075840 UP000235965 UP000075881 UP000075885 UP000007266 UP000007819 UP000015103 UP000075882 UP000075902 UP000053105 UP000183832 UP000053825 UP000008744 UP000001819 UP000192221 UP000007801 UP000000304 UP000008711 UP000268350 UP000000803 UP000002282 UP000001070 UP000092553 UP000075880 UP000005203 UP000242457 UP000008792 UP000009192 UP000009046 UP000215335 UP000198287 UP000094527 UP000007798 UP000002358 UP000008237 UP000000311 UP000076502 UP000075809 UP000078492 UP000000305 UP000005205 UP000078541 UP000007755 UP000078540 UP000076858
PRIDE
Interpro
SUPFAM
SSF50156
SSF50156
CDD
ProteinModelPortal
A0A2W1BVG6
A0A2A4JER0
A0A3S2NRV2
A0A0N0PCK6
A0A194Q6V1
A0A2H1VN75
+ More
A0A1W4WTC1 N6T1I2 A0A1Q3G0V9 Q176B0 A0A067R9K8 A0A1B6DMV8 A0A0K8TKQ3 A0A1Y1LHI3 A0A182GHT9 A0A1B6ERF9 U5ERQ4 A0A084WAJ5 A0A182M440 A0A182YFA0 A0A182RIU8 A0A182VT53 A0A182QPF2 A0A182NDD4 A0A2M3Z781 A0A2M4AJI2 W5JSW6 A0A182FTG4 A0A182VJK6 A0A182WUX1 Q7Q8X8 A0A182HYG5 A0A2J7PSG8 A0A182JQQ6 A0A182PUG8 A0A0A9YY36 A0A1L8DMQ2 A0A139WF22 A0A0T6B8A5 V5GR04 J9KAC2 A0A2S2PNE2 A0A2H8TUE7 A0A336K1L5 A0A023F4U2 A0A069DSM6 A0A0V0G9K6 T1IAA9 A0A0P4VHM2 A0A182LPX1 A0A310SIK8 A0A182TK69 A0A0M8ZRU2 A0A1J1HI01 A0A224XS57 A0A0L7RFT8 B4H9W1 Q29JD9 A0A1W4UVQ6 B3MQV2 B4R6G0 B3NWX0 A0A3B0J919 O97111 B4Q2J3 A0A1E1WPC7 A0A0A1XDA8 A0A0K8W5G0 A0A034WFY7 B4JKB8 A0A0M3QZG7 A0A0N0BDA9 W8AT66 A0A182IQJ6 A0A088A2G7 A0A2A3EB00 B4MA03 B4L7V3 E0W0G3 A0A2J7PSG9 A0A232F0U0 A0A226EX36 A0A1D2MQQ1 B4N209 K7J0F4 E2C4Z1 A0A0R3P3L1 A0A2P6L1L1 E2AIN3 A0A154NWG6 A0A151XI98 A0A151J8G3 E9FTS2 A0A158NF92 A0A151JZY1 F4WN43 A0A151I5Z4 A0A164KBQ7 A0A2H5BF90
A0A1W4WTC1 N6T1I2 A0A1Q3G0V9 Q176B0 A0A067R9K8 A0A1B6DMV8 A0A0K8TKQ3 A0A1Y1LHI3 A0A182GHT9 A0A1B6ERF9 U5ERQ4 A0A084WAJ5 A0A182M440 A0A182YFA0 A0A182RIU8 A0A182VT53 A0A182QPF2 A0A182NDD4 A0A2M3Z781 A0A2M4AJI2 W5JSW6 A0A182FTG4 A0A182VJK6 A0A182WUX1 Q7Q8X8 A0A182HYG5 A0A2J7PSG8 A0A182JQQ6 A0A182PUG8 A0A0A9YY36 A0A1L8DMQ2 A0A139WF22 A0A0T6B8A5 V5GR04 J9KAC2 A0A2S2PNE2 A0A2H8TUE7 A0A336K1L5 A0A023F4U2 A0A069DSM6 A0A0V0G9K6 T1IAA9 A0A0P4VHM2 A0A182LPX1 A0A310SIK8 A0A182TK69 A0A0M8ZRU2 A0A1J1HI01 A0A224XS57 A0A0L7RFT8 B4H9W1 Q29JD9 A0A1W4UVQ6 B3MQV2 B4R6G0 B3NWX0 A0A3B0J919 O97111 B4Q2J3 A0A1E1WPC7 A0A0A1XDA8 A0A0K8W5G0 A0A034WFY7 B4JKB8 A0A0M3QZG7 A0A0N0BDA9 W8AT66 A0A182IQJ6 A0A088A2G7 A0A2A3EB00 B4MA03 B4L7V3 E0W0G3 A0A2J7PSG9 A0A232F0U0 A0A226EX36 A0A1D2MQQ1 B4N209 K7J0F4 E2C4Z1 A0A0R3P3L1 A0A2P6L1L1 E2AIN3 A0A154NWG6 A0A151XI98 A0A151J8G3 E9FTS2 A0A158NF92 A0A151JZY1 F4WN43 A0A151I5Z4 A0A164KBQ7 A0A2H5BF90
PDB
2LC6
E-value=5.48683e-53,
Score=524
Ontologies
PATHWAY
GO
GO:0008654
GO:0016021
GO:0016780
GO:0007163
GO:0060341
GO:0017048
GO:0005634
GO:0016324
GO:0007098
GO:0005938
GO:0005080
GO:0016485
GO:0043085
GO:0097482
GO:0010592
GO:0043195
GO:0051491
GO:0006469
GO:0007430
GO:0007416
GO:0007298
GO:0045179
GO:0045200
GO:0060446
GO:0001738
GO:0035003
GO:0055059
GO:0016332
GO:0005886
GO:0045186
GO:0045176
GO:0045197
GO:0045196
GO:0005509
GO:0005515
GO:0005739
GO:0006508
GO:0008234
GO:0004890
GO:0005216
GO:0043564
GO:0006418
GO:0003824
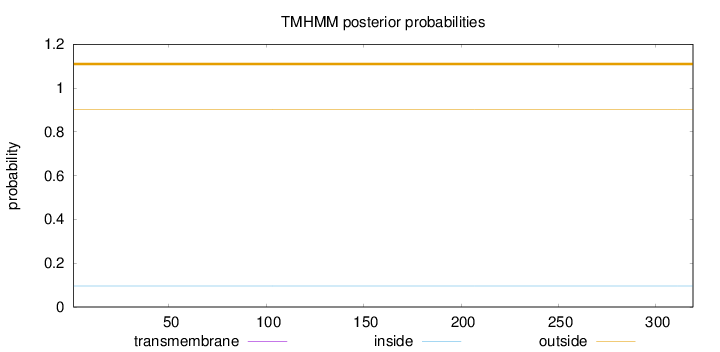
Topology
Length:
319
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00143
Exp number, first 60 AAs:
0
Total prob of N-in:
0.09645
outside
1 - 319
Population Genetic Test Statistics
Pi
147.116356
Theta
136.742709
Tajima's D
0.120258
CLR
0.675176
CSRT
0.399630018499075
Interpretation
Uncertain
