Gene
KWMTBOMO11861
Pre Gene Modal
BGIBMGA004438
Annotation
PREDICTED:_ethanolaminephosphotransferase_1-like_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 4.908
Sequence
CDS
ATGTTCAATTACAAATATTTATCACGCGAACACTTGGAGGGCTTCGACAATTACAAGTACATGGCGATCGACACGAGCCCTCTCAGTGTTTATGTCATGCACCCATTTTGGAACAAAGTTGTGGAACTTGTGCCAAAATGGATTGCACCAAATGTACTAACGTTTGCCGGATTCATGTTGGTTGTGCTCAATTTCCTGATGCTGTCATACTATGATTATGATTACTATGCTGCCAGCTTCTCAGCTAACAATGCAACATTAATTGATGCAACTACAAATGGCTATGAGGAAGTCATACCGAAATCACTGTGGTTCATAATTGCCGTATTTCTATTTCTTGCTTATACCCTAGACGGTATTGATGGGAAGCAAGCTAGACGCACCCAGACATCGGGACCGCTTGGGGAACTATTTGACCACGGTCTCGATTCATACACGGTATTCTTCATACCGGCCTGTCTATATTCTATATTTGGTCGATCGGATTACTCTATTCCGCCTATTAGGATTTATTACGTGATGTGGAATCTCTTATTGAATTTTTATTTGAGTCACTGGGAGAAATATAATACAGGGGTTCTGTTCCTGCCTTGGGGCTATGATTTTAGTATGTGGGCTTCAACCCTATGCTTCATATGGACCGGTGTTAACGGTACCTTGTTCTACAAGAAATATATCTACGGCAGTATGACGCTGGCGCACGGATTCGAACTGGCGATATACGGAACGGGAGTCGTCACCAACCTGCCAGTGGCGCTGTACAATATTAATAAGTCTTACAAGGAACGAACGGGTAAGATGCGTTCCCTCTCTGAAGCTCTAAGGCCGCTCTGGTCCTTCACATCCGTGTTCGTCATCAGCTCCGTGTGGGTGCACTGCAGCGCGCGGCTGCCGGACTACGACCTGAGGATGCTCTTCCTGTTGATCGGAACTCTGTTCAGTAACGTAGCGTGTCGTCTAATCGTCAGTCAAATGAGCAATCAACGGTGCGAAGCTATCAACTGGCTGACGTGGCCCCTGCTGGTCAGTGCTACGGTGTCATTGACACTGCCAGAGTACGAGCCGACAGCGTTCTATGGCTTCACTATGCTCACTCTATTCGCTCACATTCACTACGGAACCTGCGTTGTACGCCAGATGTGCGACCATTTCCGGGTCAGCTGTTTCCACATAAAGCAGCGTGCGGATTGA
Protein
MFNYKYLSREHLEGFDNYKYMAIDTSPLSVYVMHPFWNKVVELVPKWIAPNVLTFAGFMLVVLNFLMLSYYDYDYYAASFSANNATLIDATTNGYEEVIPKSLWFIIAVFLFLAYTLDGIDGKQARRTQTSGPLGELFDHGLDSYTVFFIPACLYSIFGRSDYSIPPIRIYYVMWNLLLNFYLSHWEKYNTGVLFLPWGYDFSMWASTLCFIWTGVNGTLFYKKYIYGSMTLAHGFELAIYGTGVVTNLPVALYNINKSYKERTGKMRSLSEALRPLWSFTSVFVISSVWVHCSARLPDYDLRMLFLLIGTLFSNVACRLIVSQMSNQRCEAINWLTWPLLVSATVSLTLPEYEPTAFYGFTMLTLFAHIHYGTCVVRQMCDHFRVSCFHIKQRAD
Summary
Similarity
Belongs to the CDP-alcohol phosphatidyltransferase class-I family.
Uniprot
H9J4J8
A0A2W1BY58
A0A2A4JE02
A0A194Q5A7
A0A0N0PCU8
A0A212FNF5
+ More
A0A2H1VN75 A0A1E1VXI1 A0A195BX17 F4WJ73 A0A195FK90 A0A158NED8 E2AHR2 A0A0L7R0J2 E2B3I2 A0A195CAF9 A0A087ZP02 A0A151J6M2 A0A026W8Y8 A0A1S3DGC7 A0A0N1PHM2 A0A194QBA1 A0A310SEQ7 A0A084VYN4 A0A154NXT5 A0A2A3ENC4 A0A182JAZ6 A0A182P677 A0A182Y934 Q17L23 A0A182F5S2 A0A1Q3F0J2 A0A1Q3F1B8 A0A1Q3F156 A0A1S4GZI5 A0A182UU48 A0A1Q3F0G2 U5EUB6 A0A182I774 Q7PWQ8 A0A182LZJ9 A0A182U723 A0A182JSQ0 A0A1B6KY01 B0WQ73 A0A182NGL6 A0A139WET6 A0A182H5V2 A0A182VUA5 A0A3L8E309 A0A182RUQ2 A0A1I8N234 A0A1B6CBA2 A0A336LNV5 A0A1L8DY92 A0A212FDP5 A0A1L8DYE3 A0A2P2I5U6 W8AMP1 A0A1I8PWB6 A0A0L0BRW0 A0A1E1W9A7 A0A0M8ZZQ1 A0A0P6I400 A0A162Q0H9 A0A224XEE3 A0A034W0B0 A0A0P5CIQ2 A0A3R7MB07 A0A0P6BQG7 A0A182RK60 A0A023F9W9 A0A1S4GZI2 A0A1A9W6V5 A0A182LDT8 A0A1A9V7B4 A0A1A9ZHB4 A0A182WFG7 A0A1S4GYS8 A0A1I8PXM5 E0VUV8 T1PBI5 T1PMW1 Q7Q8H5 B4KGK5 A0A182V8H7 A0A182KN64 A0A182IBK3 A0A1A9XCB0 A0A1D2N9D0 A0A1B0BAF5 A0A067RL51 T1PNB0 T1PL23 A0A226F5P0 A0A2M4CI30 R4G3D0 A0A2M4CID5 A0A1A9XEL3 A0A2W1BMN7
A0A2H1VN75 A0A1E1VXI1 A0A195BX17 F4WJ73 A0A195FK90 A0A158NED8 E2AHR2 A0A0L7R0J2 E2B3I2 A0A195CAF9 A0A087ZP02 A0A151J6M2 A0A026W8Y8 A0A1S3DGC7 A0A0N1PHM2 A0A194QBA1 A0A310SEQ7 A0A084VYN4 A0A154NXT5 A0A2A3ENC4 A0A182JAZ6 A0A182P677 A0A182Y934 Q17L23 A0A182F5S2 A0A1Q3F0J2 A0A1Q3F1B8 A0A1Q3F156 A0A1S4GZI5 A0A182UU48 A0A1Q3F0G2 U5EUB6 A0A182I774 Q7PWQ8 A0A182LZJ9 A0A182U723 A0A182JSQ0 A0A1B6KY01 B0WQ73 A0A182NGL6 A0A139WET6 A0A182H5V2 A0A182VUA5 A0A3L8E309 A0A182RUQ2 A0A1I8N234 A0A1B6CBA2 A0A336LNV5 A0A1L8DY92 A0A212FDP5 A0A1L8DYE3 A0A2P2I5U6 W8AMP1 A0A1I8PWB6 A0A0L0BRW0 A0A1E1W9A7 A0A0M8ZZQ1 A0A0P6I400 A0A162Q0H9 A0A224XEE3 A0A034W0B0 A0A0P5CIQ2 A0A3R7MB07 A0A0P6BQG7 A0A182RK60 A0A023F9W9 A0A1S4GZI2 A0A1A9W6V5 A0A182LDT8 A0A1A9V7B4 A0A1A9ZHB4 A0A182WFG7 A0A1S4GYS8 A0A1I8PXM5 E0VUV8 T1PBI5 T1PMW1 Q7Q8H5 B4KGK5 A0A182V8H7 A0A182KN64 A0A182IBK3 A0A1A9XCB0 A0A1D2N9D0 A0A1B0BAF5 A0A067RL51 T1PNB0 T1PL23 A0A226F5P0 A0A2M4CI30 R4G3D0 A0A2M4CID5 A0A1A9XEL3 A0A2W1BMN7
Pubmed
EMBL
BABH01021847
BABH01021848
BABH01021849
BABH01021850
KZ149924
PZC77626.1
+ More
NWSH01001730 PCG70307.1 KQ459460 KPJ00732.1 KQ460547 KPJ14042.1 AGBW02005668 OWR55268.1 ODYU01003468 SOQ42289.1 GDQN01011670 JAT79384.1 KQ976401 KYM92513.1 GL888182 EGI65651.1 KQ981490 KYN41085.1 ADTU01013259 ADTU01013260 GL439579 EFN67042.1 KQ414670 KOC64367.1 GL445346 EFN89734.1 KQ978023 KYM97859.1 KQ979851 KYN18858.1 KK107388 EZA51494.1 KQ460009 KPJ18457.1 KPJ00706.1 KQ762329 OAD55978.1 ATLV01018382 KE525231 KFB43078.1 KQ434781 KZC04486.1 KZ288204 PBC33265.1 CH477219 EAT47404.1 GFDL01013984 JAV21061.1 GFDL01013694 JAV21351.1 GFDL01013771 JAV21274.1 AAAB01008984 GFDL01014014 JAV21031.1 GANO01002397 JAB57474.1 APCN01003368 EAA14893.3 AXCM01000336 GEBQ01023650 JAT16327.1 DS232036 EDS32733.1 KQ971354 KYB26463.1 JXUM01026690 KQ560764 KXJ80976.1 QOIP01000001 RLU26765.1 GEDC01029793 GEDC01026733 GEDC01019054 GEDC01014582 GEDC01013858 GEDC01010262 GEDC01004490 GEDC01000474 JAS07505.1 JAS10565.1 JAS18244.1 JAS22716.1 JAS23440.1 JAS27036.1 JAS32808.1 JAS36824.1 UFQT01000093 SSX19610.1 GFDF01002654 JAV11430.1 AGBW02009026 OWR51833.1 GFDF01002655 JAV11429.1 IACF01003794 LAB69403.1 GAMC01019308 JAB87247.1 JRES01001455 KNC22733.1 GDQN01007501 JAT83553.1 KQ435798 KOX73386.1 GDIQ01119694 GDIQ01009909 JAN84828.1 LRGB01000389 KZS19295.1 GFTR01005589 JAW10837.1 GAKP01011227 GAKP01011226 GAKP01011225 JAC47727.1 GDIP01173682 JAJ49720.1 QCYY01002250 ROT71779.1 GDIP01011376 JAM92339.1 GBBI01000994 JAC17718.1 AAAB01008944 DS235797 EEB17164.1 KA646029 AFP60658.1 KA649425 AFP64054.1 EAA10178.4 CH933807 EDW13205.2 APCN01000708 LJIJ01000137 ODN01851.1 JXJN01010943 JXJN01010944 KK852611 KDR20264.1 KA649433 AFP64062.1 KA649426 KA649427 AFP64055.1 LNIX01000001 OXA64206.1 GGFL01000816 MBW64994.1 ACPB03011978 GAHY01001791 JAA75719.1 GGFL01000817 MBW64995.1 KZ150066 PZC74136.1
NWSH01001730 PCG70307.1 KQ459460 KPJ00732.1 KQ460547 KPJ14042.1 AGBW02005668 OWR55268.1 ODYU01003468 SOQ42289.1 GDQN01011670 JAT79384.1 KQ976401 KYM92513.1 GL888182 EGI65651.1 KQ981490 KYN41085.1 ADTU01013259 ADTU01013260 GL439579 EFN67042.1 KQ414670 KOC64367.1 GL445346 EFN89734.1 KQ978023 KYM97859.1 KQ979851 KYN18858.1 KK107388 EZA51494.1 KQ460009 KPJ18457.1 KPJ00706.1 KQ762329 OAD55978.1 ATLV01018382 KE525231 KFB43078.1 KQ434781 KZC04486.1 KZ288204 PBC33265.1 CH477219 EAT47404.1 GFDL01013984 JAV21061.1 GFDL01013694 JAV21351.1 GFDL01013771 JAV21274.1 AAAB01008984 GFDL01014014 JAV21031.1 GANO01002397 JAB57474.1 APCN01003368 EAA14893.3 AXCM01000336 GEBQ01023650 JAT16327.1 DS232036 EDS32733.1 KQ971354 KYB26463.1 JXUM01026690 KQ560764 KXJ80976.1 QOIP01000001 RLU26765.1 GEDC01029793 GEDC01026733 GEDC01019054 GEDC01014582 GEDC01013858 GEDC01010262 GEDC01004490 GEDC01000474 JAS07505.1 JAS10565.1 JAS18244.1 JAS22716.1 JAS23440.1 JAS27036.1 JAS32808.1 JAS36824.1 UFQT01000093 SSX19610.1 GFDF01002654 JAV11430.1 AGBW02009026 OWR51833.1 GFDF01002655 JAV11429.1 IACF01003794 LAB69403.1 GAMC01019308 JAB87247.1 JRES01001455 KNC22733.1 GDQN01007501 JAT83553.1 KQ435798 KOX73386.1 GDIQ01119694 GDIQ01009909 JAN84828.1 LRGB01000389 KZS19295.1 GFTR01005589 JAW10837.1 GAKP01011227 GAKP01011226 GAKP01011225 JAC47727.1 GDIP01173682 JAJ49720.1 QCYY01002250 ROT71779.1 GDIP01011376 JAM92339.1 GBBI01000994 JAC17718.1 AAAB01008944 DS235797 EEB17164.1 KA646029 AFP60658.1 KA649425 AFP64054.1 EAA10178.4 CH933807 EDW13205.2 APCN01000708 LJIJ01000137 ODN01851.1 JXJN01010943 JXJN01010944 KK852611 KDR20264.1 KA649433 AFP64062.1 KA649426 KA649427 AFP64055.1 LNIX01000001 OXA64206.1 GGFL01000816 MBW64994.1 ACPB03011978 GAHY01001791 JAA75719.1 GGFL01000817 MBW64995.1 KZ150066 PZC74136.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000078540
+ More
UP000007755 UP000078541 UP000005205 UP000000311 UP000053825 UP000008237 UP000078542 UP000005203 UP000078492 UP000053097 UP000079169 UP000030765 UP000076502 UP000242457 UP000075880 UP000075885 UP000076408 UP000008820 UP000069272 UP000075903 UP000075840 UP000007062 UP000075883 UP000075902 UP000075881 UP000002320 UP000075884 UP000007266 UP000069940 UP000249989 UP000075920 UP000279307 UP000075900 UP000095301 UP000095300 UP000037069 UP000053105 UP000076858 UP000283509 UP000091820 UP000075882 UP000078200 UP000092445 UP000009046 UP000009192 UP000092443 UP000094527 UP000092460 UP000027135 UP000198287 UP000015103
UP000007755 UP000078541 UP000005205 UP000000311 UP000053825 UP000008237 UP000078542 UP000005203 UP000078492 UP000053097 UP000079169 UP000030765 UP000076502 UP000242457 UP000075880 UP000075885 UP000076408 UP000008820 UP000069272 UP000075903 UP000075840 UP000007062 UP000075883 UP000075902 UP000075881 UP000002320 UP000075884 UP000007266 UP000069940 UP000249989 UP000075920 UP000279307 UP000075900 UP000095301 UP000095300 UP000037069 UP000053105 UP000076858 UP000283509 UP000091820 UP000075882 UP000078200 UP000092445 UP000009046 UP000009192 UP000092443 UP000094527 UP000092460 UP000027135 UP000198287 UP000015103
PRIDE
Interpro
SUPFAM
SSF50156
SSF50156
ProteinModelPortal
H9J4J8
A0A2W1BY58
A0A2A4JE02
A0A194Q5A7
A0A0N0PCU8
A0A212FNF5
+ More
A0A2H1VN75 A0A1E1VXI1 A0A195BX17 F4WJ73 A0A195FK90 A0A158NED8 E2AHR2 A0A0L7R0J2 E2B3I2 A0A195CAF9 A0A087ZP02 A0A151J6M2 A0A026W8Y8 A0A1S3DGC7 A0A0N1PHM2 A0A194QBA1 A0A310SEQ7 A0A084VYN4 A0A154NXT5 A0A2A3ENC4 A0A182JAZ6 A0A182P677 A0A182Y934 Q17L23 A0A182F5S2 A0A1Q3F0J2 A0A1Q3F1B8 A0A1Q3F156 A0A1S4GZI5 A0A182UU48 A0A1Q3F0G2 U5EUB6 A0A182I774 Q7PWQ8 A0A182LZJ9 A0A182U723 A0A182JSQ0 A0A1B6KY01 B0WQ73 A0A182NGL6 A0A139WET6 A0A182H5V2 A0A182VUA5 A0A3L8E309 A0A182RUQ2 A0A1I8N234 A0A1B6CBA2 A0A336LNV5 A0A1L8DY92 A0A212FDP5 A0A1L8DYE3 A0A2P2I5U6 W8AMP1 A0A1I8PWB6 A0A0L0BRW0 A0A1E1W9A7 A0A0M8ZZQ1 A0A0P6I400 A0A162Q0H9 A0A224XEE3 A0A034W0B0 A0A0P5CIQ2 A0A3R7MB07 A0A0P6BQG7 A0A182RK60 A0A023F9W9 A0A1S4GZI2 A0A1A9W6V5 A0A182LDT8 A0A1A9V7B4 A0A1A9ZHB4 A0A182WFG7 A0A1S4GYS8 A0A1I8PXM5 E0VUV8 T1PBI5 T1PMW1 Q7Q8H5 B4KGK5 A0A182V8H7 A0A182KN64 A0A182IBK3 A0A1A9XCB0 A0A1D2N9D0 A0A1B0BAF5 A0A067RL51 T1PNB0 T1PL23 A0A226F5P0 A0A2M4CI30 R4G3D0 A0A2M4CID5 A0A1A9XEL3 A0A2W1BMN7
A0A2H1VN75 A0A1E1VXI1 A0A195BX17 F4WJ73 A0A195FK90 A0A158NED8 E2AHR2 A0A0L7R0J2 E2B3I2 A0A195CAF9 A0A087ZP02 A0A151J6M2 A0A026W8Y8 A0A1S3DGC7 A0A0N1PHM2 A0A194QBA1 A0A310SEQ7 A0A084VYN4 A0A154NXT5 A0A2A3ENC4 A0A182JAZ6 A0A182P677 A0A182Y934 Q17L23 A0A182F5S2 A0A1Q3F0J2 A0A1Q3F1B8 A0A1Q3F156 A0A1S4GZI5 A0A182UU48 A0A1Q3F0G2 U5EUB6 A0A182I774 Q7PWQ8 A0A182LZJ9 A0A182U723 A0A182JSQ0 A0A1B6KY01 B0WQ73 A0A182NGL6 A0A139WET6 A0A182H5V2 A0A182VUA5 A0A3L8E309 A0A182RUQ2 A0A1I8N234 A0A1B6CBA2 A0A336LNV5 A0A1L8DY92 A0A212FDP5 A0A1L8DYE3 A0A2P2I5U6 W8AMP1 A0A1I8PWB6 A0A0L0BRW0 A0A1E1W9A7 A0A0M8ZZQ1 A0A0P6I400 A0A162Q0H9 A0A224XEE3 A0A034W0B0 A0A0P5CIQ2 A0A3R7MB07 A0A0P6BQG7 A0A182RK60 A0A023F9W9 A0A1S4GZI2 A0A1A9W6V5 A0A182LDT8 A0A1A9V7B4 A0A1A9ZHB4 A0A182WFG7 A0A1S4GYS8 A0A1I8PXM5 E0VUV8 T1PBI5 T1PMW1 Q7Q8H5 B4KGK5 A0A182V8H7 A0A182KN64 A0A182IBK3 A0A1A9XCB0 A0A1D2N9D0 A0A1B0BAF5 A0A067RL51 T1PNB0 T1PL23 A0A226F5P0 A0A2M4CI30 R4G3D0 A0A2M4CID5 A0A1A9XEL3 A0A2W1BMN7
Ontologies
PATHWAY
GO
PANTHER
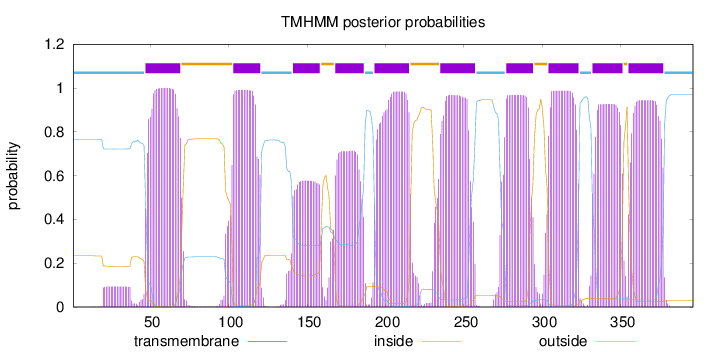
Topology
Length:
396
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
189.13624
Exp number, first 60 AAs:
14.64382
Total prob of N-in:
0.76610
POSSIBLE N-term signal
sequence
inside
1 - 46
TMhelix
47 - 69
outside
70 - 102
TMhelix
103 - 120
inside
121 - 140
TMhelix
141 - 158
outside
159 - 167
TMhelix
168 - 186
inside
187 - 192
TMhelix
193 - 215
outside
216 - 234
TMhelix
235 - 257
inside
258 - 276
TMhelix
277 - 294
outside
295 - 303
TMhelix
304 - 323
inside
324 - 331
TMhelix
332 - 351
outside
352 - 354
TMhelix
355 - 377
inside
378 - 396
Population Genetic Test Statistics
Pi
256.866687
Theta
164.224542
Tajima's D
-0.760649
CLR
16.750841
CSRT
0.184440777961102
Interpretation
Uncertain
