Gene
KWMTBOMO11857 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA004423
Annotation
PREDICTED:_protein_CASC3_isoform_X2_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.667
Sequence
CDS
ATGGTTTTAGCGTCCCAAAAAACACCAAACACCTTCTTGCGTTTCCATCCAAAAGTTGTACACTGTAGGCACAAGCTCATTCAAAATGCCGCCAATCTGGAAGTTATACAAGTGACACGGGAGGACGCCGAAATGACGTCCGTGGTACGAAGACGGGAGCACGACGATTCGGGAGAATATTCAGATGCTTCTCAGGACATCGAACACAATAACTCGTTGAACGAAGGGCCGCCCGAGTCCGATTATGACTCGCAGGGTAGCGATTCTGACCACTCTGAACGGGATCCAGAGGCACAGGAAGGTGAACGCAGAGTTGATGATGATGAAGATAGGAGCAATCCTCAGTATATCCCCAAGAGAGGAACATTCTACGAGCATGATGACCGAACCGCAGCAAGCGGCGAGGAATCTGTCAACACGACATCCGAAACGGCTTCAGAGAAAAAGGAGGGCTCCGAGACGACGACCGAACGACGCCGCCCTCCACGAAAATCTGACATTGTCAATAAATGGACGCACGACAAATACAACGAGAATGAACAGATGCCGAAATCACGTGACGAGCTGGTAGCGATCTACGGGTACGACATACGCAACGAAGATGCCCCTCCTCACGCGAGACGCAACAGAAGATACGGTCGTGGACCCAACAAGTACACACGTACCTGGGAAGACGAGGAGGCGTACCGAAGACAGAACGCGTTGCCGCTGAGAAAACCACCGAACCCCGAGGACTTCCCTGAACTTGAAGCTAGCGCTAAGAAACCAGGGAGGAAGGTTCGCGCCCCCCGGCCTCGGTCCAAGTCGCAGCCGGCGGCGGGCGAGTCCCCGGAGCGCGCCGCCCAGCTCCGCCGGAACGCCTCCATCAAAACCGCGAAGCCCAAGCCGCCGTTCACAAAGACGACGCCCCCGAAGAACCTGAAGCCGCAGGACAAGAAATTACCGGCGCTCGAAACTTTGACAATCACCGCTCCGGCGAATAATAACAATAACAACAATAACAGCCAAGGTCAGTCGCAGCGGAAGGTGAGCGCTATGGTGGCGCAGCAGCGGCCAGTGCCCACGGACATCAACAAGAAGAAGCCGCTTCCCGGCCAGAACCAGCAGCCCTCGAGTCCAGCCCCCAACAAGTCGCCGCCCCTGCAAACGAAACCACAGAACCAGACCTCTCACCCTCACCCGCAACAGACCACTGGCGCCGAGCCCCGCGGCGGCAGCAAGCGCTACAGCAGCCTGCGGCAGCGCCCCCTCGCTGAGCCCTACACGCCCCAACCGCAGCCGCAGACGCAGACCCAGACCCCTGTCCAGACCTCTAGTCCGGGGGCAGCGCAGACCCAGCAACCGCACAGATATGCCGGTCAAAGCGAGTTCACCGCCGGCCCGGCCGCGCCTGCACCCCACGCGCCGCACCTCGCGCACCAGGTGCGGTGCACGCCGAACGCGCCGGCCCCCGGACACACGCCGCCGCCCCTCCAGGCTCTGCCACAACAACAACAACAACAGCAGCAGCAGGTCAGACTGGGGATCATTGATCAGCCGCAACAAGTCGTGTCACATCCCCACCACCCCATACACAACATACAGCCGCAGCCTCATACGATAAATCAATTACCACCCGCCCAGGGATACGCGCCCCCCGCCATGATGCCCGCGCAGTACATGCAGCAGAGCGGCGCGGCGCCCGTGTTCGGCGGCGGGCTGTTCCCGCAGGGCGCGGCGCCCCCCTACCCGCTCTACCCGCACCACAACTACGCGACCCAAACCGGTGGGGTGACGTATTACAACTGCGCGGAGCAGGACCAACAGCCGCGTGCCCAGCGGCGCCCCACCGCAGCCATACCCATCGTCAAGCCGCACGCCTCTGAAAGAGCTGCCAATTCAACCGAGAAAGACAACATAGATCGTATCGTCGAGAATATGTTCGTGAGGAAGCCCTGGCCCGCGACGCACGGAGCCGATAAAGAGAAATCATCGGAGGCCCGGAGTTCGCAAGAGTCCGGCAGCAAGGAGTCCTCGCAGCCCGACAGCCTCACGTCCAGCTCCATCTCGACGGACTCGAGACCTTCGGAGGCCAAGGAAGAGAAAGAGAAGGACAGCCGAGAGAACGAGACGGAGCAACCGCCCGAGGCCTGA
Protein
MVLASQKTPNTFLRFHPKVVHCRHKLIQNAANLEVIQVTREDAEMTSVVRRREHDDSGEYSDASQDIEHNNSLNEGPPESDYDSQGSDSDHSERDPEAQEGERRVDDDEDRSNPQYIPKRGTFYEHDDRTAASGEESVNTTSETASEKKEGSETTTERRRPPRKSDIVNKWTHDKYNENEQMPKSRDELVAIYGYDIRNEDAPPHARRNRRYGRGPNKYTRTWEDEEAYRRQNALPLRKPPNPEDFPELEASAKKPGRKVRAPRPRSKSQPAAGESPERAAQLRRNASIKTAKPKPPFTKTTPPKNLKPQDKKLPALETLTITAPANNNNNNNNSQGQSQRKVSAMVAQQRPVPTDINKKKPLPGQNQQPSSPAPNKSPPLQTKPQNQTSHPHPQQTTGAEPRGGSKRYSSLRQRPLAEPYTPQPQPQTQTQTPVQTSSPGAAQTQQPHRYAGQSEFTAGPAAPAPHAPHLAHQVRCTPNAPAPGHTPPPLQALPQQQQQQQQQVRLGIIDQPQQVVSHPHHPIHNIQPQPHTINQLPPAQGYAPPAMMPAQYMQQSGAAPVFGGGLFPQGAAPPYPLYPHHNYATQTGGVTYYNCAEQDQQPRAQRRPTAAIPIVKPHASERAANSTEKDNIDRIVENMFVRKPWPATHGADKEKSSEARSSQESGSKESSQPDSLTSSSISTDSRPSEAKEEKEKDSRENETEQPPEA
Summary
Uniprot
Pubmed
EMBL
Pfam
PF09405 Btz
ProteinModelPortal
PDB
2XB2
E-value=8.96479e-07,
Score=129
Ontologies
PATHWAY
PANTHER
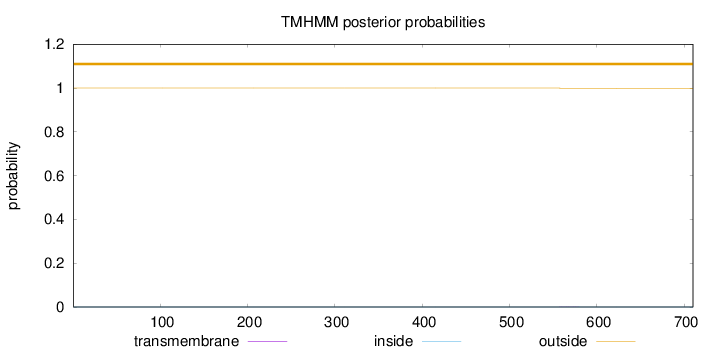
Topology
Length:
710
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00558
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00020
outside
1 - 710
Population Genetic Test Statistics
Pi
206.617644
Theta
164.950217
Tajima's D
-1.324559
CLR
1830.164728
CSRT
0.0837958102094895
Interpretation
Possibly Positive selection
