Gene
KWMTBOMO11854
Pre Gene Modal
BGIBMGA004424
Annotation
neuropeptide_receptor_A16_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.83
Sequence
CDS
ATGGCCGTGTCATTTGTGAATAAATTATTTTTATTTATCTTTGTGCTAAACAAAGAAAATATATTAGCAGACGATGATGTCACAACCAAACAATTGAATTTGTACATAGCTGACAAACAAGAATCTTTTGAAATGACAACTGTAGAAGATGATCTGAATGTACCAATAAAAATGAAAGCCAACAAAATCATAAGCGAATATGATGATAGATTTAAAACAGATACGAACTCTAGTGAATTTGAGGAAGCAGAGAATGAAACCTGCGTTGGAGATCCACAATACTGCAACATGACAAAGGAGGAGTATGTCAAAATGATACAAGAATATATTTACCCAAACCCGTACGAGTGGATATTGATAGCTACACACACGTTTGTGTTCATAACCGGATTATTTGGGAACGCGCTGGTGTGCGTCGCGGTTTACAGGAATCACTCAATGCGGACTGTTACGAATTATTTCATTGTCAACCTAGCTGTGGCTGATTTCATGGTGATATTGTTTTGTCTGCCAGCGACGGTGCTGTGGGACGTGACTGAGACTTGGTTCCTTGGGGAAGGGCTTTGTAAAGTCCTGCCCTATTTTCAGTCAGTGTCAGTGACGGTGTCAGTTCTGACGCTGACTTTCATCTCTGTGGACCGCTGGTACGCTATCTGCTTCCCGCTGAAGTTCAAGTCGACCACGGGCCGCGCCAAGACCGCCATACTGATTATATGGCTCGTCTCTTTGTGCTTCAATATACCGGAGCTAGTGGTGCTGAAGCTGGTCCGATTCGTTCCTCTGCGCTTCGAGCTGCCGTACTTGCTGCAATGCTACGGGACCTGGTCTCCGAGCAGCGAACTCGTTTGGCACATCCTCAAAGTCCTTTTGATATATACACTACCCCTCGTGCTCATGGCCGTCGCGTATCATCAGATAGCGAGGGTGCTTTGGAGCTCCAACGGAATTCCCGGACAGGCCGACACAAAGAAACTGGCCACTGCAGAACTGAAACACGGCTATAAGATTCTATTTCGATTATACAGGGTGTTCCACAAACATTATATCAAGCCAAAAGCTGTAAATAGTGTTCTGTTAGAATAA
Protein
MAVSFVNKLFLFIFVLNKENILADDDVTTKQLNLYIADKQESFEMTTVEDDLNVPIKMKANKIISEYDDRFKTDTNSSEFEEAENETCVGDPQYCNMTKEEYVKMIQEYIYPNPYEWILIATHTFVFITGLFGNALVCVAVYRNHSMRTVTNYFIVNLAVADFMVILFCLPATVLWDVTETWFLGEGLCKVLPYFQSVSVTVSVLTLTFISVDRWYAICFPLKFKSTTGRAKTAILIIWLVSLCFNIPELVVLKLVRFVPLRFELPYLLQCYGTWSPSSELVWHILKVLLIYTLPLVLMAVAYHQIARVLWSSNGIPGQADTKKLATAELKHGYKILFRLYRVFHKHYIKPKAVNSVLLE
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family.
Uniprot
B3XXM9
A0A0S1YD80
A0A345ANM7
A0A0E3EKU4
F1CZQ1
A0A0N1PJF5
+ More
A0A212FAI2 A0A0N0PF28 A0A0S1YD65 B3XXL7 A0A2A4J7H0 A0A1B6C133 A0A1B6KTL7 I6NNS5 A0A336M9M0 A0A1B6JDZ3 U3U4E1 A0A2S2PRZ9 A0A182Q9T1 A0A2C9GPU2 A0A1J1HTG2 G3JWZ1 A0A310SW43 A0A087ZYQ8 K7IWF9 A0A2U9PG22 A0A151X4S1 A0A158NR92 A0A0C9R4T1 A0A195FJP6 A0A151ITZ3 A0A1Y1MF02 A0A0P6B7K0 A0A0A9WR33 A0A0A9WMC0 A0A0P5W420 A0A026W5R0 T1IYS5 A0A1S3D076 A0A2C9JHH0 W5LZB7 A0A3L8DIJ3 A0A0K0PVS5
A0A212FAI2 A0A0N0PF28 A0A0S1YD65 B3XXL7 A0A2A4J7H0 A0A1B6C133 A0A1B6KTL7 I6NNS5 A0A336M9M0 A0A1B6JDZ3 U3U4E1 A0A2S2PRZ9 A0A182Q9T1 A0A2C9GPU2 A0A1J1HTG2 G3JWZ1 A0A310SW43 A0A087ZYQ8 K7IWF9 A0A2U9PG22 A0A151X4S1 A0A158NR92 A0A0C9R4T1 A0A195FJP6 A0A151ITZ3 A0A1Y1MF02 A0A0P6B7K0 A0A0A9WR33 A0A0A9WMC0 A0A0P5W420 A0A026W5R0 T1IYS5 A0A1S3D076 A0A2C9JHH0 W5LZB7 A0A3L8DIJ3 A0A0K0PVS5
Pubmed
EMBL
AB330437
BAG68415.1
KT031014
ALM88312.1
MF770987
AXF38049.1
+ More
KF881015 AIT70966.1 HQ634154 ADX66344.1 KQ459718 KPJ20251.1 AGBW02009467 OWR50755.1 LADI01012895 KPJ20751.1 KT031002 ALM88300.1 AB330425 BAG68403.1 NWSH01002529 PCG68067.1 GEDC01030319 JAS06979.1 GEBQ01025181 JAT14796.1 JN543509 AEX08666.2 UFQS01000581 UFQT01000581 SSX05122.1 SSX25483.1 GECU01010359 JAS97347.1 AB817299 BAO01066.1 GGMR01019621 MBY32240.1 AXCN02001908 AXCN02001909 APCN01005316 CVRI01000020 CRK91360.1 JN030894 AEN03789.1 KQ759870 OAD62416.1 MG550205 AWT50639.1 KQ982540 KYQ55426.1 ADTU01023826 ADTU01023827 ADTU01023828 GBYB01002980 GBYB01002983 JAG72747.1 JAG72750.1 KQ981522 KYN40586.1 KQ980970 KYN10929.1 GEZM01033168 JAV84382.1 GDIP01020859 JAM82856.1 GBHO01034662 GBHO01034661 GDHC01019905 JAG08942.1 JAG08943.1 JAP98723.1 GBHO01034664 GBHO01034660 GDHC01012203 GDHC01004088 JAG08940.1 JAG08944.1 JAQ06426.1 JAQ14541.1 GDIP01091981 JAM11734.1 KK107390 EZA51412.1 JH431696 AHAT01009366 AHAT01009367 QOIP01000007 RLU20260.1 KP294022 AKQ63076.1
KF881015 AIT70966.1 HQ634154 ADX66344.1 KQ459718 KPJ20251.1 AGBW02009467 OWR50755.1 LADI01012895 KPJ20751.1 KT031002 ALM88300.1 AB330425 BAG68403.1 NWSH01002529 PCG68067.1 GEDC01030319 JAS06979.1 GEBQ01025181 JAT14796.1 JN543509 AEX08666.2 UFQS01000581 UFQT01000581 SSX05122.1 SSX25483.1 GECU01010359 JAS97347.1 AB817299 BAO01066.1 GGMR01019621 MBY32240.1 AXCN02001908 AXCN02001909 APCN01005316 CVRI01000020 CRK91360.1 JN030894 AEN03789.1 KQ759870 OAD62416.1 MG550205 AWT50639.1 KQ982540 KYQ55426.1 ADTU01023826 ADTU01023827 ADTU01023828 GBYB01002980 GBYB01002983 JAG72747.1 JAG72750.1 KQ981522 KYN40586.1 KQ980970 KYN10929.1 GEZM01033168 JAV84382.1 GDIP01020859 JAM82856.1 GBHO01034662 GBHO01034661 GDHC01019905 JAG08942.1 JAG08943.1 JAP98723.1 GBHO01034664 GBHO01034660 GDHC01012203 GDHC01004088 JAG08940.1 JAG08944.1 JAQ06426.1 JAQ14541.1 GDIP01091981 JAM11734.1 KK107390 EZA51412.1 JH431696 AHAT01009366 AHAT01009367 QOIP01000007 RLU20260.1 KP294022 AKQ63076.1
Proteomes
Pfam
PF00001 7tm_1
Interpro
CDD
ProteinModelPortal
B3XXM9
A0A0S1YD80
A0A345ANM7
A0A0E3EKU4
F1CZQ1
A0A0N1PJF5
+ More
A0A212FAI2 A0A0N0PF28 A0A0S1YD65 B3XXL7 A0A2A4J7H0 A0A1B6C133 A0A1B6KTL7 I6NNS5 A0A336M9M0 A0A1B6JDZ3 U3U4E1 A0A2S2PRZ9 A0A182Q9T1 A0A2C9GPU2 A0A1J1HTG2 G3JWZ1 A0A310SW43 A0A087ZYQ8 K7IWF9 A0A2U9PG22 A0A151X4S1 A0A158NR92 A0A0C9R4T1 A0A195FJP6 A0A151ITZ3 A0A1Y1MF02 A0A0P6B7K0 A0A0A9WR33 A0A0A9WMC0 A0A0P5W420 A0A026W5R0 T1IYS5 A0A1S3D076 A0A2C9JHH0 W5LZB7 A0A3L8DIJ3 A0A0K0PVS5
A0A212FAI2 A0A0N0PF28 A0A0S1YD65 B3XXL7 A0A2A4J7H0 A0A1B6C133 A0A1B6KTL7 I6NNS5 A0A336M9M0 A0A1B6JDZ3 U3U4E1 A0A2S2PRZ9 A0A182Q9T1 A0A2C9GPU2 A0A1J1HTG2 G3JWZ1 A0A310SW43 A0A087ZYQ8 K7IWF9 A0A2U9PG22 A0A151X4S1 A0A158NR92 A0A0C9R4T1 A0A195FJP6 A0A151ITZ3 A0A1Y1MF02 A0A0P6B7K0 A0A0A9WR33 A0A0A9WMC0 A0A0P5W420 A0A026W5R0 T1IYS5 A0A1S3D076 A0A2C9JHH0 W5LZB7 A0A3L8DIJ3 A0A0K0PVS5
PDB
5WS3
E-value=1.43149e-53,
Score=529
Ontologies
GO
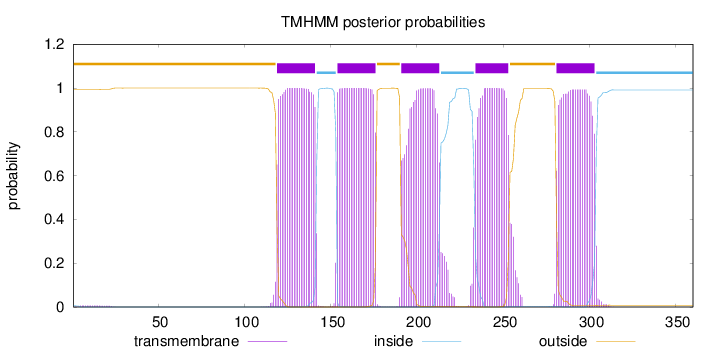
Topology
Length:
360
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
111.74348
Exp number, first 60 AAs:
0.11631
Total prob of N-in:
0.00644
outside
1 - 118
TMhelix
119 - 141
inside
142 - 153
TMhelix
154 - 176
outside
177 - 190
TMhelix
191 - 213
inside
214 - 233
TMhelix
234 - 253
outside
254 - 280
TMhelix
281 - 303
inside
304 - 360
Population Genetic Test Statistics
Pi
214.026644
Theta
169.544725
Tajima's D
1.493168
CLR
0.270693
CSRT
0.787060646967652
Interpretation
Uncertain
