Gene
KWMTBOMO11851
Pre Gene Modal
BGIBMGA004425
Annotation
Trypsin_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 0.97
Sequence
CDS
ATGAATATCTACATTTGCGTATTATTATTTATTAAAACTGTATCGAGTAAGATAGAAACGGTACAGCGACTTAGCAATGCTGAATTAGATGAGCACACGAGAGTGACGAACAGTCACGTATTCCCGTACGTGGTTGCGATACTGCAGCGATCGAAGTACGTCAGCGCGGGGGCCTTGATTGACGAGAATTGGATTTTGACGGCCGCCGACGGATTGTTCCTAATGAGAGAAACTTTAAAACTAGTGAAAGTGAGACTCGGGAGTATCAATTACAAAAAAGGAGGGCTGCTACTGCCGGCCAAGCTGATACAAATTCATCCTTATTTCGACGACAAAAGTCCCATTTTCGATGTGGCCATGGTCCAGCTGGCTGAAACCCTGAGGTTCACGCCGAAAATAAAAGCGATAAGACTTCAGAAGACGTTCATGGATGTTGCCGCAACTCATTTCATTGTGACGTCATGGAATCCATCGTTGAAACAAAAGCCCAAACACCCCGAATCGATGGAAGCTATAGAGCGCCGCCGAATGCTCACAGTGACTCATCTCCATCCATCGGAAGTTGAGGAGTGCGCCGCCGAGCTGGACGCCTACGGTGTCAATAAGACTGACTTGATCATGTGTCTGGATCCTCCTGGTGGAAGTGAAATTTGCGTGGAAAATCGGAACGTGGGTGCTCCAGTAGTGCTCAATGGAGTTCTCTGGGGTGTAGTGTCGTCTTGGAAGACAAGTGATTGTGACCAAGGGGCAGTCGGACCTAGTTTCGTGACCAGAGTGTCGGCACCTGACGTTACATCTTGGATACACGCCACATTGCACGGACATCGGTGGAAGCATAAATCGAACGAATAG
Protein
MNIYICVLLFIKTVSSKIETVQRLSNAELDEHTRVTNSHVFPYVVAILQRSKYVSAGALIDENWILTAADGLFLMRETLKLVKVRLGSINYKKGGLLLPAKLIQIHPYFDDKSPIFDVAMVQLAETLRFTPKIKAIRLQKTFMDVAATHFIVTSWNPSLKQKPKHPESMEAIERRRMLTVTHLHPSEVEECAAELDAYGVNKTDLIMCLDPPGGSEICVENRNVGAPVVLNGVLWGVVSSWKTSDCDQGAVGPSFVTRVSAPDVTSWIHATLHGHRWKHKSNE
Summary
Similarity
Belongs to the peptidase S1 family.
Uniprot
H9J4I5
A0A2W1BWP4
A0A2A4IWZ1
A0A2H1W522
A0A212FAI6
A0A0N1IH57
+ More
A0A0N1PIR5 A0A194Q5E2 A0A0N1IQB1 A0A2A4JYQ1 A0A2H1W6G0 A0A3S2L7J7 A0A0L7L4N4 A0A158NXQ1 A0A195B099 A0A2P8YZF7 A0A023ENK6 A0A195EUU2 A0A151IPG0 A0A1A9VTQ1 A0A195EDZ4 Q29JR8 A0A3B0JXQ2 B4GJ91 A0A182Q1U0 A0A023EM39 B4JQ66 F4WTJ3 A0A1W4XBM5 A0A1A9Z6S9 A0A0J9QZW1 A0A1A9W0A9 Q9VLF5 B3MUR5 B3DMZ7 A0A1B6KWQ6 A0A182MLX4 Q5QBG0 Q24091 A0A023ENI8 A0A182K9Z7 A0A023EMF3 A0A3S2TDP7 B8XY09 F4MI42 B0XLQ8 B4HYU1 A0A1Y1LWY0 O18600 A0A151WQ85 B5AKB2 A0A0L7L3T3 D3TMZ8 A0A2H1W7U3 B0WE94 B4LUZ8 A0A023EMG0 A0A182VQI7 D3TL15 A0A1B0G1B6 A0A182RWW9 Q56GM3 A0A2H1VRF0 A0A1B0AYX9 L0ATN6 Q7Q290 A0A182PUR2 B4NY05 A0A182XH38 A0A182KMS5 A0A182I8B4 A0A182VCG5 A0A1B0B1H8 A0A0L1J2D2 B0W771 B3N7W6 B0WDC9 Q7Q2Q8 A0A087QK59 A0A1B0F938 A0A1V1FT97 A0A182WYK2 E1ZYQ4 A0A3G5BIL4 A0A1Q3FTA7 A0A182U656 A0A1B0G3Q9 A0A0K8TYY4 A0A182XXN7 A0A1W4XBV3 A0A1I8N7Q1 A0A1A9XEJ7 Q5QBG3 A0A2A4JXN3 A0A091V3L3 A0A084VY00 A0A1W4WDS1 B0WUK3
A0A0N1PIR5 A0A194Q5E2 A0A0N1IQB1 A0A2A4JYQ1 A0A2H1W6G0 A0A3S2L7J7 A0A0L7L4N4 A0A158NXQ1 A0A195B099 A0A2P8YZF7 A0A023ENK6 A0A195EUU2 A0A151IPG0 A0A1A9VTQ1 A0A195EDZ4 Q29JR8 A0A3B0JXQ2 B4GJ91 A0A182Q1U0 A0A023EM39 B4JQ66 F4WTJ3 A0A1W4XBM5 A0A1A9Z6S9 A0A0J9QZW1 A0A1A9W0A9 Q9VLF5 B3MUR5 B3DMZ7 A0A1B6KWQ6 A0A182MLX4 Q5QBG0 Q24091 A0A023ENI8 A0A182K9Z7 A0A023EMF3 A0A3S2TDP7 B8XY09 F4MI42 B0XLQ8 B4HYU1 A0A1Y1LWY0 O18600 A0A151WQ85 B5AKB2 A0A0L7L3T3 D3TMZ8 A0A2H1W7U3 B0WE94 B4LUZ8 A0A023EMG0 A0A182VQI7 D3TL15 A0A1B0G1B6 A0A182RWW9 Q56GM3 A0A2H1VRF0 A0A1B0AYX9 L0ATN6 Q7Q290 A0A182PUR2 B4NY05 A0A182XH38 A0A182KMS5 A0A182I8B4 A0A182VCG5 A0A1B0B1H8 A0A0L1J2D2 B0W771 B3N7W6 B0WDC9 Q7Q2Q8 A0A087QK59 A0A1B0F938 A0A1V1FT97 A0A182WYK2 E1ZYQ4 A0A3G5BIL4 A0A1Q3FTA7 A0A182U656 A0A1B0G3Q9 A0A0K8TYY4 A0A182XXN7 A0A1W4XBV3 A0A1I8N7Q1 A0A1A9XEJ7 Q5QBG3 A0A2A4JXN3 A0A091V3L3 A0A084VY00 A0A1W4WDS1 B0WUK3
Pubmed
19121390
28756777
22118469
26354079
26227816
21347285
+ More
29403074 24945155 15632085 17994087 23185243 21719571 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 8682311 21569254 28004739 20353571 16247003 12364791 14747013 17210077 17550304 20966253 28410430 20798317 30400621 25244985 25315136 15796745 24438588
29403074 24945155 15632085 17994087 23185243 21719571 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 8682311 21569254 28004739 20353571 16247003 12364791 14747013 17210077 17550304 20966253 28410430 20798317 30400621 25244985 25315136 15796745 24438588
EMBL
BABH01021817
BABH01021818
KZ149924
PZC77617.1
NWSH01005274
PCG64325.1
+ More
ODYU01006381 SOQ48179.1 AGBW02009467 OWR50754.1 KQ459718 KPJ20252.1 LADI01012895 KPJ20753.1 KQ459460 KPJ00763.1 KQ459838 KPJ19753.1 NWSH01000350 PCG77151.1 ODYU01006643 SOQ48670.1 RSAL01001561 RVE40760.1 JTDY01002946 KOB70432.1 ADTU01003291 KQ976692 KYM77715.1 PYGN01000271 PSN49618.1 GAPW01003098 JAC10500.1 KQ981958 KYN32013.1 KQ976864 KYN07850.1 KQ979074 KYN23022.1 CH379062 EAL32893.3 KRT05049.1 KRT05050.1 KRT05051.1 OUUW01000010 SPP85843.1 CH479184 EDW37405.1 AXCN02000286 GAPW01003247 JAC10351.1 CH916372 EDV99046.1 GL888336 EGI62511.1 CM002910 KMY89204.1 AE014134 AAF52738.1 AHN54286.1 CH902624 EDV32980.1 BT032785 BT032800 ACD81799.1 ACD81814.1 GEBQ01024112 JAT15865.1 AXCM01000261 AY752851 AAV84264.1 U28641 AAC47304.1 GAPW01003097 JAC10501.1 GAPW01003100 JAC10498.1 RSAL01000281 RVE43032.1 FJ458410 ACL37992.1 EZ933290 ADJ57671.1 DS234650 EDS34693.1 CH480818 EDW52221.1 GEZM01044920 JAV78042.1 U93213 AAB66371.1 KQ982851 KYQ49855.1 EU855138 ACF72874.2 JTDY01003193 KOB69986.1 EZ422800 ADD19076.1 ODYU01006890 SOQ49140.1 DS231906 EDS45388.1 CH940649 EDW63247.1 GAPW01003096 JAC10502.1 EZ422106 ADD18310.1 CCAG010004614 AY958426 AAX59050.1 ODYU01003960 SOQ43356.1 JXJN01006043 JX915894 AFZ78860.1 AAAB01008978 EAA13597.5 CM000157 EDW88607.2 APCN01002693 JXJN01007177 JNOM01000151 KNG85578.1 DS231852 EDS37676.1 CH954177 EDV58327.1 DS231896 EDS44442.1 AAAB01008968 EAA13158.3 KL225709 KFM01613.1 FX985332 BAX07345.1 GL435242 EFN73594.1 MK075220 AYV99623.1 GFDL01004392 JAV30653.1 CCAG010009053 GDHF01032625 JAI19689.1 AY752848 AAV84261.1 NWSH01000434 PCG76448.1 KL410524 KFQ96915.1 ATLV01018234 KE525226 KFB42844.1 DS232107 EDS34988.1
ODYU01006381 SOQ48179.1 AGBW02009467 OWR50754.1 KQ459718 KPJ20252.1 LADI01012895 KPJ20753.1 KQ459460 KPJ00763.1 KQ459838 KPJ19753.1 NWSH01000350 PCG77151.1 ODYU01006643 SOQ48670.1 RSAL01001561 RVE40760.1 JTDY01002946 KOB70432.1 ADTU01003291 KQ976692 KYM77715.1 PYGN01000271 PSN49618.1 GAPW01003098 JAC10500.1 KQ981958 KYN32013.1 KQ976864 KYN07850.1 KQ979074 KYN23022.1 CH379062 EAL32893.3 KRT05049.1 KRT05050.1 KRT05051.1 OUUW01000010 SPP85843.1 CH479184 EDW37405.1 AXCN02000286 GAPW01003247 JAC10351.1 CH916372 EDV99046.1 GL888336 EGI62511.1 CM002910 KMY89204.1 AE014134 AAF52738.1 AHN54286.1 CH902624 EDV32980.1 BT032785 BT032800 ACD81799.1 ACD81814.1 GEBQ01024112 JAT15865.1 AXCM01000261 AY752851 AAV84264.1 U28641 AAC47304.1 GAPW01003097 JAC10501.1 GAPW01003100 JAC10498.1 RSAL01000281 RVE43032.1 FJ458410 ACL37992.1 EZ933290 ADJ57671.1 DS234650 EDS34693.1 CH480818 EDW52221.1 GEZM01044920 JAV78042.1 U93213 AAB66371.1 KQ982851 KYQ49855.1 EU855138 ACF72874.2 JTDY01003193 KOB69986.1 EZ422800 ADD19076.1 ODYU01006890 SOQ49140.1 DS231906 EDS45388.1 CH940649 EDW63247.1 GAPW01003096 JAC10502.1 EZ422106 ADD18310.1 CCAG010004614 AY958426 AAX59050.1 ODYU01003960 SOQ43356.1 JXJN01006043 JX915894 AFZ78860.1 AAAB01008978 EAA13597.5 CM000157 EDW88607.2 APCN01002693 JXJN01007177 JNOM01000151 KNG85578.1 DS231852 EDS37676.1 CH954177 EDV58327.1 DS231896 EDS44442.1 AAAB01008968 EAA13158.3 KL225709 KFM01613.1 FX985332 BAX07345.1 GL435242 EFN73594.1 MK075220 AYV99623.1 GFDL01004392 JAV30653.1 CCAG010009053 GDHF01032625 JAI19689.1 AY752848 AAV84261.1 NWSH01000434 PCG76448.1 KL410524 KFQ96915.1 ATLV01018234 KE525226 KFB42844.1 DS232107 EDS34988.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000283053
+ More
UP000037510 UP000005205 UP000078540 UP000245037 UP000078541 UP000078542 UP000078200 UP000078492 UP000001819 UP000268350 UP000008744 UP000075886 UP000001070 UP000007755 UP000192223 UP000092445 UP000091820 UP000000803 UP000007801 UP000075883 UP000075881 UP000002320 UP000001292 UP000075809 UP000008792 UP000075920 UP000092444 UP000075900 UP000092460 UP000007062 UP000075885 UP000002282 UP000076407 UP000075882 UP000075840 UP000075903 UP000037505 UP000008711 UP000053286 UP000092443 UP000000311 UP000075902 UP000076408 UP000095301 UP000053283 UP000030765 UP000192221
UP000037510 UP000005205 UP000078540 UP000245037 UP000078541 UP000078542 UP000078200 UP000078492 UP000001819 UP000268350 UP000008744 UP000075886 UP000001070 UP000007755 UP000192223 UP000092445 UP000091820 UP000000803 UP000007801 UP000075883 UP000075881 UP000002320 UP000001292 UP000075809 UP000008792 UP000075920 UP000092444 UP000075900 UP000092460 UP000007062 UP000075885 UP000002282 UP000076407 UP000075882 UP000075840 UP000075903 UP000037505 UP000008711 UP000053286 UP000092443 UP000000311 UP000075902 UP000076408 UP000095301 UP000053283 UP000030765 UP000192221
Pfam
PF00089 Trypsin
Interpro
SUPFAM
SSF50494
SSF50494
CDD
ProteinModelPortal
H9J4I5
A0A2W1BWP4
A0A2A4IWZ1
A0A2H1W522
A0A212FAI6
A0A0N1IH57
+ More
A0A0N1PIR5 A0A194Q5E2 A0A0N1IQB1 A0A2A4JYQ1 A0A2H1W6G0 A0A3S2L7J7 A0A0L7L4N4 A0A158NXQ1 A0A195B099 A0A2P8YZF7 A0A023ENK6 A0A195EUU2 A0A151IPG0 A0A1A9VTQ1 A0A195EDZ4 Q29JR8 A0A3B0JXQ2 B4GJ91 A0A182Q1U0 A0A023EM39 B4JQ66 F4WTJ3 A0A1W4XBM5 A0A1A9Z6S9 A0A0J9QZW1 A0A1A9W0A9 Q9VLF5 B3MUR5 B3DMZ7 A0A1B6KWQ6 A0A182MLX4 Q5QBG0 Q24091 A0A023ENI8 A0A182K9Z7 A0A023EMF3 A0A3S2TDP7 B8XY09 F4MI42 B0XLQ8 B4HYU1 A0A1Y1LWY0 O18600 A0A151WQ85 B5AKB2 A0A0L7L3T3 D3TMZ8 A0A2H1W7U3 B0WE94 B4LUZ8 A0A023EMG0 A0A182VQI7 D3TL15 A0A1B0G1B6 A0A182RWW9 Q56GM3 A0A2H1VRF0 A0A1B0AYX9 L0ATN6 Q7Q290 A0A182PUR2 B4NY05 A0A182XH38 A0A182KMS5 A0A182I8B4 A0A182VCG5 A0A1B0B1H8 A0A0L1J2D2 B0W771 B3N7W6 B0WDC9 Q7Q2Q8 A0A087QK59 A0A1B0F938 A0A1V1FT97 A0A182WYK2 E1ZYQ4 A0A3G5BIL4 A0A1Q3FTA7 A0A182U656 A0A1B0G3Q9 A0A0K8TYY4 A0A182XXN7 A0A1W4XBV3 A0A1I8N7Q1 A0A1A9XEJ7 Q5QBG3 A0A2A4JXN3 A0A091V3L3 A0A084VY00 A0A1W4WDS1 B0WUK3
A0A0N1PIR5 A0A194Q5E2 A0A0N1IQB1 A0A2A4JYQ1 A0A2H1W6G0 A0A3S2L7J7 A0A0L7L4N4 A0A158NXQ1 A0A195B099 A0A2P8YZF7 A0A023ENK6 A0A195EUU2 A0A151IPG0 A0A1A9VTQ1 A0A195EDZ4 Q29JR8 A0A3B0JXQ2 B4GJ91 A0A182Q1U0 A0A023EM39 B4JQ66 F4WTJ3 A0A1W4XBM5 A0A1A9Z6S9 A0A0J9QZW1 A0A1A9W0A9 Q9VLF5 B3MUR5 B3DMZ7 A0A1B6KWQ6 A0A182MLX4 Q5QBG0 Q24091 A0A023ENI8 A0A182K9Z7 A0A023EMF3 A0A3S2TDP7 B8XY09 F4MI42 B0XLQ8 B4HYU1 A0A1Y1LWY0 O18600 A0A151WQ85 B5AKB2 A0A0L7L3T3 D3TMZ8 A0A2H1W7U3 B0WE94 B4LUZ8 A0A023EMG0 A0A182VQI7 D3TL15 A0A1B0G1B6 A0A182RWW9 Q56GM3 A0A2H1VRF0 A0A1B0AYX9 L0ATN6 Q7Q290 A0A182PUR2 B4NY05 A0A182XH38 A0A182KMS5 A0A182I8B4 A0A182VCG5 A0A1B0B1H8 A0A0L1J2D2 B0W771 B3N7W6 B0WDC9 Q7Q2Q8 A0A087QK59 A0A1B0F938 A0A1V1FT97 A0A182WYK2 E1ZYQ4 A0A3G5BIL4 A0A1Q3FTA7 A0A182U656 A0A1B0G3Q9 A0A0K8TYY4 A0A182XXN7 A0A1W4XBV3 A0A1I8N7Q1 A0A1A9XEJ7 Q5QBG3 A0A2A4JXN3 A0A091V3L3 A0A084VY00 A0A1W4WDS1 B0WUK3
PDB
1SLW
E-value=6.35422e-10,
Score=152
Ontologies
GO
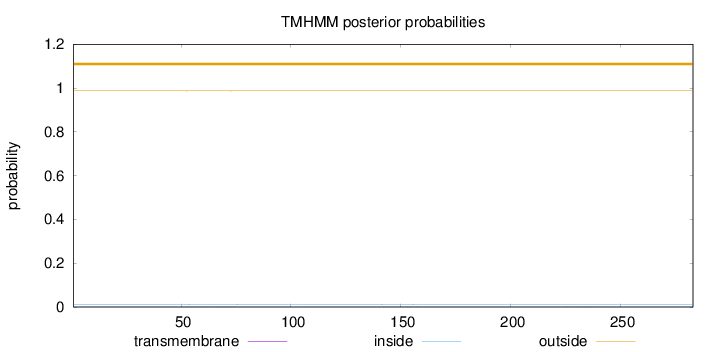
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.827141
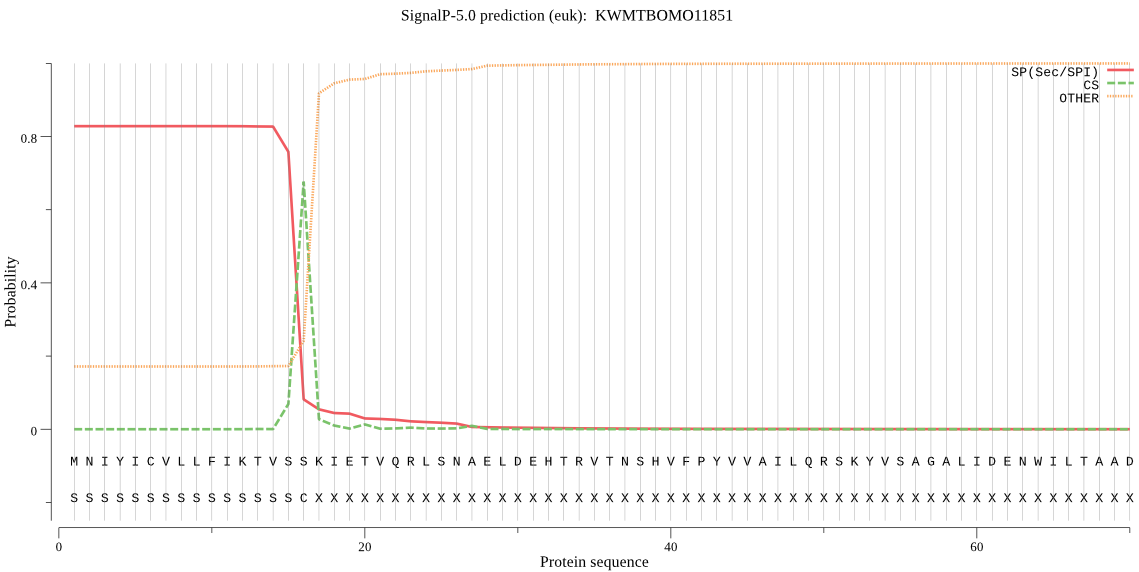
Length:
283
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0248599999999999
Exp number, first 60 AAs:
0.0061
Total prob of N-in:
0.01268
outside
1 - 283
Population Genetic Test Statistics
Pi
155.529624
Theta
30.055489
Tajima's D
0.981298
CLR
19.138625
CSRT
0.654767261636918
Interpretation
Uncertain
