Gene
KWMTBOMO11850
Pre Gene Modal
BGIBMGA004426
Annotation
putative_sugar_transporter_[Danaus_plexippus]
Location in the cell
PlasmaMembrane Reliability : 4.586
Sequence
CDS
ATGGTTTACACGGTTAATTGTGATCGGGTTGTGATCACCAAGGATGTTGAACGTGGCAGTGTATGGCGCGGCGTGGTCATAGCCCTCATCGCAAGCTTAGGATTCTTCACGCACGGCATTCAAACTGCCAATTTGACGTCCACAGCCCATTCGGGGCACTTCATCAACACAGACCACGTACCTTGGAGTACAACTGCTCTAGTGATCACAGCAGCCATAGCCGGTCCAGTGTTTTGTTTCACCATCGACAGACATGGAAGGAAAATGGGAATTTTTATTATTAATCTGGTTCAAGGTGCTTCCTTAATACCCTTGTTCTTTCTAAACGATACAAGCACAATCATTCTACACGTAATCGCCGGCATGGCCACTGGAGGCTTATTCACCGTATGCCCAATATACATCCAAGAAATATCATCGCTAAAAACAAAAGGCTTCTCAATAGAGGATGTTCTTTATGGTGGCATTGGTGATGTTCCAGTTTATTTTGATGGTCTTCGTATTGGAGTCTCCTAG
Protein
MVYTVNCDRVVITKDVERGSVWRGVVIALIASLGFFTHGIQTANLTSTAHSGHFINTDHVPWSTTALVITAAIAGPVFCFTIDRHGRKMGIFIINLVQGASLIPLFFLNDTSTIILHVIAGMATGGLFTVCPIYIQEISSLKTKGFSIEDVLYGGIGDVPVYFDGLRIGVS
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9J4I6
A0A2W1BWR3
A0A2A4K2C2
A0A212EWK9
A0A2H1W5S2
A0A2A4K2Q1
+ More
A0A0N1PK00 A0A212F0M2 H9JFD3 A0A2H1WM77 A0A2A4JKE4 A0A212EZE7 A0A2H1X046 A0A2A4J9N1 A0A0L7L9U3 A0A2A4JNL8 A0A3S2NZV3 A0A1B6C0L1 A0A336LM27 A0A1V8SKU6 A0A0L7L525 A0A1B6M474 H9JVT8 A0A3S1B0L8 A0A2P7YG71 A0A2W1BTH5 A0A194QBF3 A0A161MCD9 A0A1V8TJ41 A0A0K8WHX8 A0A1B6JHT2 A0A319DEF6 A0A1B6HTA4 A0A1Y2HBC3 A0A3S2WFA8 A0A292PPD6 A0A364KXV2 W3X680 A0A1V8TRZ7 A0A1L7XUS3 A0A1V8ULU9 B8MIV7 A0A034W3I3 M7SL55 B4NLR2 A0A366S9Y6 A0A317VAM5 A0A0B7NUN8 A0A1Q8S606 A0A2C7Z0P9 A0A3N5YVU5 A0A1L7VEM7 A0A365NJ65 A0A0I9YBH4 A0A1V6QJ80 A0A1L7VZW7 D7GGZ8 A0A2C8AXC9 A0A225AVM5 S0EDM6 A0A2H3SFC8 A0A160VK86 A0A0A8Q552 A0A2C8AFQ9 A0A2C7ZRR5 A0A365MY23 A0A2D3USS0 A0A0M8PC89 A5Y0C3 A0A164I902 A0A117NMB6 A0A1V6T274 A0A074W283
A0A0N1PK00 A0A212F0M2 H9JFD3 A0A2H1WM77 A0A2A4JKE4 A0A212EZE7 A0A2H1X046 A0A2A4J9N1 A0A0L7L9U3 A0A2A4JNL8 A0A3S2NZV3 A0A1B6C0L1 A0A336LM27 A0A1V8SKU6 A0A0L7L525 A0A1B6M474 H9JVT8 A0A3S1B0L8 A0A2P7YG71 A0A2W1BTH5 A0A194QBF3 A0A161MCD9 A0A1V8TJ41 A0A0K8WHX8 A0A1B6JHT2 A0A319DEF6 A0A1B6HTA4 A0A1Y2HBC3 A0A3S2WFA8 A0A292PPD6 A0A364KXV2 W3X680 A0A1V8TRZ7 A0A1L7XUS3 A0A1V8ULU9 B8MIV7 A0A034W3I3 M7SL55 B4NLR2 A0A366S9Y6 A0A317VAM5 A0A0B7NUN8 A0A1Q8S606 A0A2C7Z0P9 A0A3N5YVU5 A0A1L7VEM7 A0A365NJ65 A0A0I9YBH4 A0A1V6QJ80 A0A1L7VZW7 D7GGZ8 A0A2C8AXC9 A0A225AVM5 S0EDM6 A0A2H3SFC8 A0A160VK86 A0A0A8Q552 A0A2C8AFQ9 A0A2C7ZRR5 A0A365MY23 A0A2D3USS0 A0A0M8PC89 A5Y0C3 A0A164I902 A0A117NMB6 A0A1V6T274 A0A074W283
Pubmed
EMBL
BABH01021813
KZ149924
PZC77637.1
NWSH01000245
PCG78054.1
AGBW02011935
+ More
OWR45888.1 ODYU01006208 SOQ47854.1 PCG78053.1 KQ459718 KPJ20253.1 AGBW02011050 OWR47308.1 BABH01038297 ODYU01009532 SOQ54032.1 NWSH01001270 PCG71903.1 AGBW02011317 OWR46831.1 ODYU01012350 SOQ58622.1 NWSH01002244 PCG68837.1 JTDY01002032 KOB72273.1 NWSH01001033 PCG73010.1 RSAL01000077 RVE48769.1 GEDC01030246 JAS07052.1 UFQT01000043 SSX18775.1 NAJO01000038 OQN99699.1 JTDY01002946 KOB70431.1 GEBQ01009252 JAT30725.1 BABH01040665 BABH01040666 BABH01040667 RQTK01001132 RUS71996.1 NHZQ01000445 PSK34965.1 KZ149954 PZC76507.1 KQ459460 KPJ00761.1 GEMB01001415 JAS01741.1 NAJO01000007 OQO11386.1 GDHF01001576 JAI50738.1 GECU01008989 JAS98717.1 KZ825848 PYH95736.1 GECU01029810 JAS77896.1 MCFL01000054 ORZ31878.1 RZYA01000014 RVU20979.1 LN891138 CUS08350.1 MIKG01000007 RAO68364.1 KI912112 ETS81549.1 NAJO01000002 OQO13982.1 FJOG01000059 CZR68779.1 NAEU01000333 OQO24397.1 EQ962657 EED15619.1 GAKP01009698 JAC49254.1 KB706967 EMR64937.1 CH964274 EDW85301.1 QKXC01000027 RBR26143.1 MSFL01000030 PWY70429.1 LM676409 CEP26406.1 MPGH01000014 OLN96842.1 LT618779 LT618780 SCQ47156.1 SCQ50628.1 RPON01001400 RPJ77722.1 FJOF01000003 CZR37895.1 PKMI01000008 RBA20588.1 JRVG01000067 KLO92540.1 MDYO01000064 OQD89299.1 FJOF01000010 CZR45959.1 FN806773 CBL57809.1 LT599498 LT618789 LT618790 SBW75733.1 SCQ65591.1 SCQ75320.1 LFMY01000009 OKL58485.1 HF679030 CCT71947.1 FMJU01000009 SCO10756.1 LN997841 CUW08218.1 CDAH01000042 LT618776 LT618777 CEI24112.1 SCQ45034.1 SCQ48723.1 LT618791 LT618792 SCQ69760.1 SCQ78795.1 LT618781 LT618785 LT618786 LT618782 SCQ57976.1 SCQ59299.1 SCQ65907.1 SCQ66179.1 PKMI01000028 RBA13453.1 FJUY01000003 CZT16865.1 LHQQ01000048 KOS45161.1 EF028238 ABM01870.1 LWGR01000021 KZM69211.1 LLXE01000250 KUM58971.1 MLKD01000013 OQE20332.1 KL584825 KEQ66913.1
OWR45888.1 ODYU01006208 SOQ47854.1 PCG78053.1 KQ459718 KPJ20253.1 AGBW02011050 OWR47308.1 BABH01038297 ODYU01009532 SOQ54032.1 NWSH01001270 PCG71903.1 AGBW02011317 OWR46831.1 ODYU01012350 SOQ58622.1 NWSH01002244 PCG68837.1 JTDY01002032 KOB72273.1 NWSH01001033 PCG73010.1 RSAL01000077 RVE48769.1 GEDC01030246 JAS07052.1 UFQT01000043 SSX18775.1 NAJO01000038 OQN99699.1 JTDY01002946 KOB70431.1 GEBQ01009252 JAT30725.1 BABH01040665 BABH01040666 BABH01040667 RQTK01001132 RUS71996.1 NHZQ01000445 PSK34965.1 KZ149954 PZC76507.1 KQ459460 KPJ00761.1 GEMB01001415 JAS01741.1 NAJO01000007 OQO11386.1 GDHF01001576 JAI50738.1 GECU01008989 JAS98717.1 KZ825848 PYH95736.1 GECU01029810 JAS77896.1 MCFL01000054 ORZ31878.1 RZYA01000014 RVU20979.1 LN891138 CUS08350.1 MIKG01000007 RAO68364.1 KI912112 ETS81549.1 NAJO01000002 OQO13982.1 FJOG01000059 CZR68779.1 NAEU01000333 OQO24397.1 EQ962657 EED15619.1 GAKP01009698 JAC49254.1 KB706967 EMR64937.1 CH964274 EDW85301.1 QKXC01000027 RBR26143.1 MSFL01000030 PWY70429.1 LM676409 CEP26406.1 MPGH01000014 OLN96842.1 LT618779 LT618780 SCQ47156.1 SCQ50628.1 RPON01001400 RPJ77722.1 FJOF01000003 CZR37895.1 PKMI01000008 RBA20588.1 JRVG01000067 KLO92540.1 MDYO01000064 OQD89299.1 FJOF01000010 CZR45959.1 FN806773 CBL57809.1 LT599498 LT618789 LT618790 SBW75733.1 SCQ65591.1 SCQ75320.1 LFMY01000009 OKL58485.1 HF679030 CCT71947.1 FMJU01000009 SCO10756.1 LN997841 CUW08218.1 CDAH01000042 LT618776 LT618777 CEI24112.1 SCQ45034.1 SCQ48723.1 LT618791 LT618792 SCQ69760.1 SCQ78795.1 LT618781 LT618785 LT618786 LT618782 SCQ57976.1 SCQ59299.1 SCQ65907.1 SCQ66179.1 PKMI01000028 RBA13453.1 FJUY01000003 CZT16865.1 LHQQ01000048 KOS45161.1 EF028238 ABM01870.1 LWGR01000021 KZM69211.1 LLXE01000250 KUM58971.1 MLKD01000013 OQE20332.1 KL584825 KEQ66913.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000037510
UP000283053
+ More
UP000192596 UP000271974 UP000243723 UP000053268 UP000247810 UP000193411 UP000283128 UP000249363 UP000030651 UP000184330 UP000192386 UP000001745 UP000012174 UP000007798 UP000253153 UP000247233 UP000186583 UP000245659 UP000183971 UP000251714 UP000036287 UP000191612 UP000000936 UP000220705 UP000245389 UP000214365 UP000016800 UP000219504 UP000076747 UP000049868 UP000250020 UP000245943 UP000245888 UP000246141 UP000225277 UP000037696 UP000076512 UP000055045 UP000191285 UP000030672
UP000192596 UP000271974 UP000243723 UP000053268 UP000247810 UP000193411 UP000283128 UP000249363 UP000030651 UP000184330 UP000192386 UP000001745 UP000012174 UP000007798 UP000253153 UP000247233 UP000186583 UP000245659 UP000183971 UP000251714 UP000036287 UP000191612 UP000000936 UP000220705 UP000245389 UP000214365 UP000016800 UP000219504 UP000076747 UP000049868 UP000250020 UP000245943 UP000245888 UP000246141 UP000225277 UP000037696 UP000076512 UP000055045 UP000191285 UP000030672
Interpro
ProteinModelPortal
H9J4I6
A0A2W1BWR3
A0A2A4K2C2
A0A212EWK9
A0A2H1W5S2
A0A2A4K2Q1
+ More
A0A0N1PK00 A0A212F0M2 H9JFD3 A0A2H1WM77 A0A2A4JKE4 A0A212EZE7 A0A2H1X046 A0A2A4J9N1 A0A0L7L9U3 A0A2A4JNL8 A0A3S2NZV3 A0A1B6C0L1 A0A336LM27 A0A1V8SKU6 A0A0L7L525 A0A1B6M474 H9JVT8 A0A3S1B0L8 A0A2P7YG71 A0A2W1BTH5 A0A194QBF3 A0A161MCD9 A0A1V8TJ41 A0A0K8WHX8 A0A1B6JHT2 A0A319DEF6 A0A1B6HTA4 A0A1Y2HBC3 A0A3S2WFA8 A0A292PPD6 A0A364KXV2 W3X680 A0A1V8TRZ7 A0A1L7XUS3 A0A1V8ULU9 B8MIV7 A0A034W3I3 M7SL55 B4NLR2 A0A366S9Y6 A0A317VAM5 A0A0B7NUN8 A0A1Q8S606 A0A2C7Z0P9 A0A3N5YVU5 A0A1L7VEM7 A0A365NJ65 A0A0I9YBH4 A0A1V6QJ80 A0A1L7VZW7 D7GGZ8 A0A2C8AXC9 A0A225AVM5 S0EDM6 A0A2H3SFC8 A0A160VK86 A0A0A8Q552 A0A2C8AFQ9 A0A2C7ZRR5 A0A365MY23 A0A2D3USS0 A0A0M8PC89 A5Y0C3 A0A164I902 A0A117NMB6 A0A1V6T274 A0A074W283
A0A0N1PK00 A0A212F0M2 H9JFD3 A0A2H1WM77 A0A2A4JKE4 A0A212EZE7 A0A2H1X046 A0A2A4J9N1 A0A0L7L9U3 A0A2A4JNL8 A0A3S2NZV3 A0A1B6C0L1 A0A336LM27 A0A1V8SKU6 A0A0L7L525 A0A1B6M474 H9JVT8 A0A3S1B0L8 A0A2P7YG71 A0A2W1BTH5 A0A194QBF3 A0A161MCD9 A0A1V8TJ41 A0A0K8WHX8 A0A1B6JHT2 A0A319DEF6 A0A1B6HTA4 A0A1Y2HBC3 A0A3S2WFA8 A0A292PPD6 A0A364KXV2 W3X680 A0A1V8TRZ7 A0A1L7XUS3 A0A1V8ULU9 B8MIV7 A0A034W3I3 M7SL55 B4NLR2 A0A366S9Y6 A0A317VAM5 A0A0B7NUN8 A0A1Q8S606 A0A2C7Z0P9 A0A3N5YVU5 A0A1L7VEM7 A0A365NJ65 A0A0I9YBH4 A0A1V6QJ80 A0A1L7VZW7 D7GGZ8 A0A2C8AXC9 A0A225AVM5 S0EDM6 A0A2H3SFC8 A0A160VK86 A0A0A8Q552 A0A2C8AFQ9 A0A2C7ZRR5 A0A365MY23 A0A2D3USS0 A0A0M8PC89 A5Y0C3 A0A164I902 A0A117NMB6 A0A1V6T274 A0A074W283
PDB
6H7D
E-value=0.0848534,
Score=78
Ontologies
GO
PANTHER
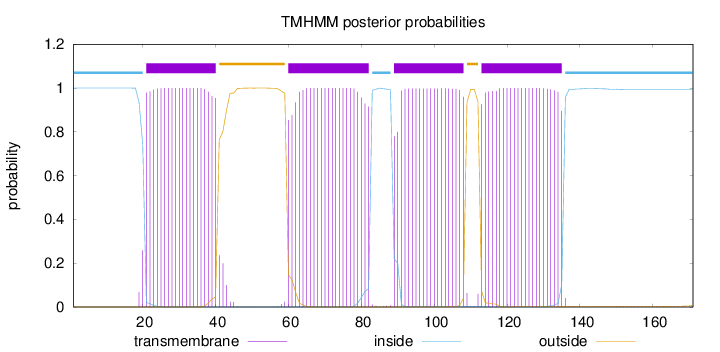
Topology
Length:
171
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
85.71741
Exp number, first 60 AAs:
21.65177
Total prob of N-in:
0.99989
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 40
outside
41 - 59
TMhelix
60 - 82
inside
83 - 88
TMhelix
89 - 108
outside
109 - 112
TMhelix
113 - 135
inside
136 - 171
Population Genetic Test Statistics
Pi
248.192636
Theta
155.86263
Tajima's D
1.867748
CLR
0.168445
CSRT
0.860956952152392
Interpretation
Uncertain
