Gene
KWMTBOMO11844
Pre Gene Modal
BGIBMGA004429
Annotation
neuropeptide_receptor_A5_precursor_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.959
Sequence
CDS
ATGGCACTTAGAAAAGAATCCTTGGCCATAATCACGATGTTGATCATATGCAATTACGTCCTAAGTTCAAACTTCGATTCGATACCAGAGAGCATCAGAGTAAGAAAGTCCGTTGATAACACCACTTCGAGGAGTAGCCTAAAAAATTTAAACGAGACAATGAAACAAAGCAATAATGAAACTGAATTTGGAAGGCTTCTAGATGCAACCGAAATGACAACTGAATATGATAATTTCACTGAAGAGCCCTGCGTTGGCGACCGGGCCTTCTGCAATTTGACCAGAGAGGAGTACATGGAGATGCTGAATGATTACGTGTTCCCTCAGCCCTACGAATGGGTACTCATAGCTACTCACGCCATAGTATTTGTGATCGGGCTGATAGGGAATGCGCTTGTCTGCATAGCGGTGTATAGAAATCATTCCATGAGAACTGTCACGAATTATTTCATCGTGAATCTGGCTGTGGCCGATTTTATGGTGATATTGATATGTCTGCCGCCGACTGTGCTCTGGGACGTCACCGAAACCTGGTTCTTTGGGACAGCCATGTGCAGGATCGTCCTGTACTTTCAGTCAGTGTCAGTGACGGTGTCAGTTCTGACGCTGACGTTCATCTCTGTGGACCGCTGGTACGCTATCTGCTTCCCGCTGAAGTTCAAGTCAACCACGGGCCGCGCCAAGACCGCTATACTGATTATATGGCTTCTATCTCTACTATTCAATATACCGGAATTCGTGGTGCTTCAAGTGCAAACGAAGATGCAGCTGCGCTTCAACGTGCAATACTTCATGCAGTGCGCTTCCACTTGGTCTGATGAAAGCGATCTGACTTGGCACATCATAAAGGCACTGTTTCTGTATACATTCCCGCTATTGCTGATGACGATCGCGTACTGTCAAATCGTACGAGTCCTTTGGAGATCAGACAACATTCCTGGCCACACTGAATCCCACAAGCTGTGCAGCACGCAAACAGGGCAGAGCAATTGGCTGGCGGCATCTCGACGCACAACGCCATCGATCCACACGAACGCCTCCACCGAGGGTCAGCTTCGGTCACGGCGGAAGGCCGCCAAGATGCTGGTGGCTGTGGTAGCGATGTTCGCTGTTTGTTACTTCCCGGTACACCTCCTCTCTGTATTAAGGGTCGCTTTTGACGTGCAACAAACAGACGTGATGACCTGCATCGCTCTGATATCACACGTGATGTGTTACGCGAACTCCGCCGTCAACCCGCTGATTTACAACTTCATGAGCGGGAAATTCCGTCGCGAATTCCACAGATCGTACTTCAAATGTTTCTGCTGCTGTCACACCACGCCGGCGCCAGAACAGAATGGGGCTTCGTTTGAGCCAATCGGCAGTTCTAGAGCTCGGACCATCCGCACCACGGTCCGACGGCACGACTCCTGCGTCAGCTATCGACTCGCCCATCTGTCACCGTCGAACCACAACATTCACCGGGACTACATTCAAAATACGAACACGTCTTTTATCGAGCCAATGAACGGAAATCGCAGGTCTAAAATCAGGGACGAATCGATCAGCGACACCGCGACACGTTTCACGGTGACCACCGATATCCCTTGTAAAGATTGA
Protein
MALRKESLAIITMLIICNYVLSSNFDSIPESIRVRKSVDNTTSRSSLKNLNETMKQSNNETEFGRLLDATEMTTEYDNFTEEPCVGDRAFCNLTREEYMEMLNDYVFPQPYEWVLIATHAIVFVIGLIGNALVCIAVYRNHSMRTVTNYFIVNLAVADFMVILICLPPTVLWDVTETWFFGTAMCRIVLYFQSVSVTVSVLTLTFISVDRWYAICFPLKFKSTTGRAKTAILIIWLLSLLFNIPEFVVLQVQTKMQLRFNVQYFMQCASTWSDESDLTWHIIKALFLYTFPLLLMTIAYCQIVRVLWRSDNIPGHTESHKLCSTQTGQSNWLAASRRTTPSIHTNASTEGQLRSRRKAAKMLVAVVAMFAVCYFPVHLLSVLRVAFDVQQTDVMTCIALISHVMCYANSAVNPLIYNFMSGKFRREFHRSYFKCFCCCHTTPAPEQNGASFEPIGSSRARTIRTTVRRHDSCVSYRLAHLSPSNHNIHRDYIQNTNTSFIEPMNGNRRSKIRDESISDTATRFTVTTDIPCKD
Summary
Similarity
Belongs to the G-protein coupled receptor 1 family.
Uniprot
B3XXL7
A0A0S1YD65
A0A0N0PF28
A0A0N1IB59
A0A2A4J7H0
A0A0E3EKU4
+ More
A0A345ANM7 F1CZQ1 A0A0S1YD80 B3XXM9 A0A212FAI2 G3JWZ1 U5N0U6 A0A1B6KTL7 U3U4E1 A0A1B6JDZ3 A0A2C9GPU2 A0A1B6C133 A0A1S4GM37 I6NNS5 A0A310SW43 K7IWF9 A0A154PAQ2 A0A0A9WMC0 A0A087ZYQ8 A0A0A9WR33 A0A2U9PG22 A0A0P5AJC7 A0A0P5NAX8 A0A195FJP6 A0A0N8DCT1 A0A0N8BU42 A0A0P5KQM3 A0A162P288 A0A0P6F5P7 A0A0P5T2I6 A0A0C9R4T1 A0A026W5R0 A0A151ITZ3 A0A1W4XMD0 A0A195BFF0 A0A1S3D076 A0A2I7NB16 A0A0K0PUH3 A0A0K0PVS5 A0A1W0WY80 R7VBE1 A0A2C9JHH0
A0A345ANM7 F1CZQ1 A0A0S1YD80 B3XXM9 A0A212FAI2 G3JWZ1 U5N0U6 A0A1B6KTL7 U3U4E1 A0A1B6JDZ3 A0A2C9GPU2 A0A1B6C133 A0A1S4GM37 I6NNS5 A0A310SW43 K7IWF9 A0A154PAQ2 A0A0A9WMC0 A0A087ZYQ8 A0A0A9WR33 A0A2U9PG22 A0A0P5AJC7 A0A0P5NAX8 A0A195FJP6 A0A0N8DCT1 A0A0N8BU42 A0A0P5KQM3 A0A162P288 A0A0P6F5P7 A0A0P5T2I6 A0A0C9R4T1 A0A026W5R0 A0A151ITZ3 A0A1W4XMD0 A0A195BFF0 A0A1S3D076 A0A2I7NB16 A0A0K0PUH3 A0A0K0PVS5 A0A1W0WY80 R7VBE1 A0A2C9JHH0
Pubmed
EMBL
AB330425
BAG68403.1
KT031002
ALM88300.1
LADI01012895
KPJ20751.1
+ More
KQ459718 KPJ20254.1 NWSH01002529 PCG68067.1 KF881015 AIT70966.1 MF770987 AXF38049.1 HQ634154 ADX66344.1 KT031014 ALM88312.1 AB330437 BAG68415.1 AGBW02009467 OWR50755.1 JN030894 AEN03789.1 KC439534 AGX85002.1 GEBQ01025181 JAT14796.1 AB817299 BAO01066.1 GECU01010359 JAS97347.1 APCN01005316 GEDC01030319 JAS06979.1 AAAB01008968 JN543509 AEX08666.2 KQ759870 OAD62416.1 KQ434864 KZC09016.1 GBHO01034664 GBHO01034660 GDHC01012203 GDHC01004088 JAG08940.1 JAG08944.1 JAQ06426.1 JAQ14541.1 GBHO01034662 GBHO01034661 GDHC01019905 JAG08942.1 JAG08943.1 JAP98723.1 MG550205 AWT50639.1 GDIP01198480 JAJ24922.1 GDIQ01144722 JAL07004.1 KQ981522 KYN40586.1 GDIP01046399 JAM57316.1 GDIQ01144723 JAL07003.1 GDIQ01182063 JAK69662.1 LRGB01000512 KZS18457.1 GDIQ01065110 JAN29627.1 GDIQ01097150 JAL54576.1 GBYB01002980 GBYB01002983 JAG72747.1 JAG72750.1 KK107390 EZA51412.1 KQ980970 KYN10929.1 KQ976504 KYM82895.1 MF467262 AUR53643.1 KP293976 AKQ63030.1 KP294022 AKQ63076.1 MTYJ01000032 OQV20160.1 AMQN01004356 KB293301 ELU16138.1
KQ459718 KPJ20254.1 NWSH01002529 PCG68067.1 KF881015 AIT70966.1 MF770987 AXF38049.1 HQ634154 ADX66344.1 KT031014 ALM88312.1 AB330437 BAG68415.1 AGBW02009467 OWR50755.1 JN030894 AEN03789.1 KC439534 AGX85002.1 GEBQ01025181 JAT14796.1 AB817299 BAO01066.1 GECU01010359 JAS97347.1 APCN01005316 GEDC01030319 JAS06979.1 AAAB01008968 JN543509 AEX08666.2 KQ759870 OAD62416.1 KQ434864 KZC09016.1 GBHO01034664 GBHO01034660 GDHC01012203 GDHC01004088 JAG08940.1 JAG08944.1 JAQ06426.1 JAQ14541.1 GBHO01034662 GBHO01034661 GDHC01019905 JAG08942.1 JAG08943.1 JAP98723.1 MG550205 AWT50639.1 GDIP01198480 JAJ24922.1 GDIQ01144722 JAL07004.1 KQ981522 KYN40586.1 GDIP01046399 JAM57316.1 GDIQ01144723 JAL07003.1 GDIQ01182063 JAK69662.1 LRGB01000512 KZS18457.1 GDIQ01065110 JAN29627.1 GDIQ01097150 JAL54576.1 GBYB01002980 GBYB01002983 JAG72747.1 JAG72750.1 KK107390 EZA51412.1 KQ980970 KYN10929.1 KQ976504 KYM82895.1 MF467262 AUR53643.1 KP293976 AKQ63030.1 KP294022 AKQ63076.1 MTYJ01000032 OQV20160.1 AMQN01004356 KB293301 ELU16138.1
Proteomes
Pfam
PF00001 7tm_1
Interpro
CDD
ProteinModelPortal
B3XXL7
A0A0S1YD65
A0A0N0PF28
A0A0N1IB59
A0A2A4J7H0
A0A0E3EKU4
+ More
A0A345ANM7 F1CZQ1 A0A0S1YD80 B3XXM9 A0A212FAI2 G3JWZ1 U5N0U6 A0A1B6KTL7 U3U4E1 A0A1B6JDZ3 A0A2C9GPU2 A0A1B6C133 A0A1S4GM37 I6NNS5 A0A310SW43 K7IWF9 A0A154PAQ2 A0A0A9WMC0 A0A087ZYQ8 A0A0A9WR33 A0A2U9PG22 A0A0P5AJC7 A0A0P5NAX8 A0A195FJP6 A0A0N8DCT1 A0A0N8BU42 A0A0P5KQM3 A0A162P288 A0A0P6F5P7 A0A0P5T2I6 A0A0C9R4T1 A0A026W5R0 A0A151ITZ3 A0A1W4XMD0 A0A195BFF0 A0A1S3D076 A0A2I7NB16 A0A0K0PUH3 A0A0K0PVS5 A0A1W0WY80 R7VBE1 A0A2C9JHH0
A0A345ANM7 F1CZQ1 A0A0S1YD80 B3XXM9 A0A212FAI2 G3JWZ1 U5N0U6 A0A1B6KTL7 U3U4E1 A0A1B6JDZ3 A0A2C9GPU2 A0A1B6C133 A0A1S4GM37 I6NNS5 A0A310SW43 K7IWF9 A0A154PAQ2 A0A0A9WMC0 A0A087ZYQ8 A0A0A9WR33 A0A2U9PG22 A0A0P5AJC7 A0A0P5NAX8 A0A195FJP6 A0A0N8DCT1 A0A0N8BU42 A0A0P5KQM3 A0A162P288 A0A0P6F5P7 A0A0P5T2I6 A0A0C9R4T1 A0A026W5R0 A0A151ITZ3 A0A1W4XMD0 A0A195BFF0 A0A1S3D076 A0A2I7NB16 A0A0K0PUH3 A0A0K0PVS5 A0A1W0WY80 R7VBE1 A0A2C9JHH0
PDB
5WS3
E-value=3.24933e-54,
Score=537
Ontologies
KEGG
GO
Topology
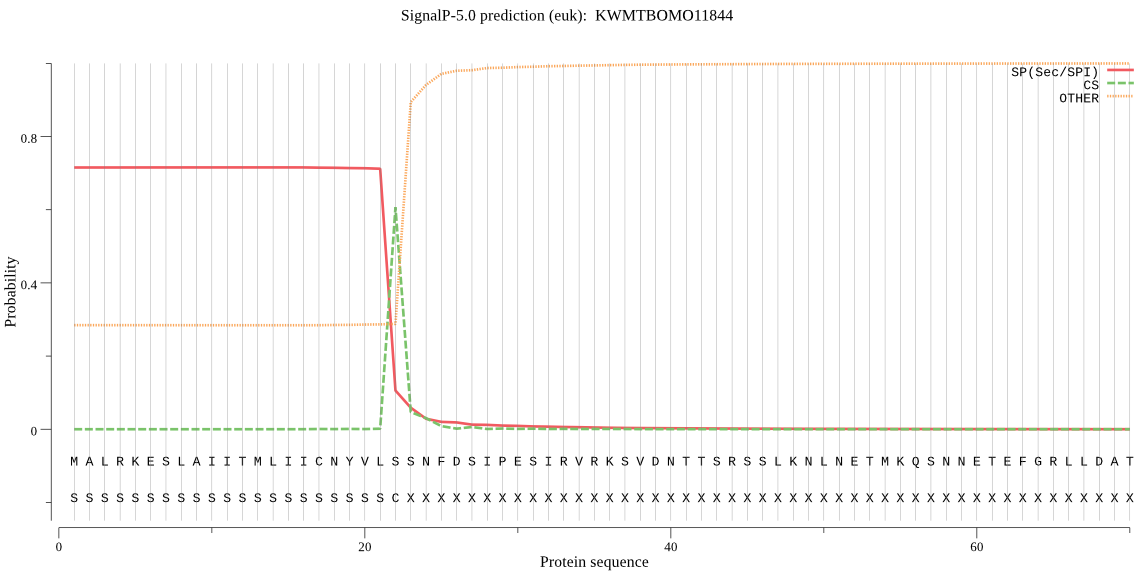
SignalP
Position: 1 - 22,
Likelihood: 0.715154
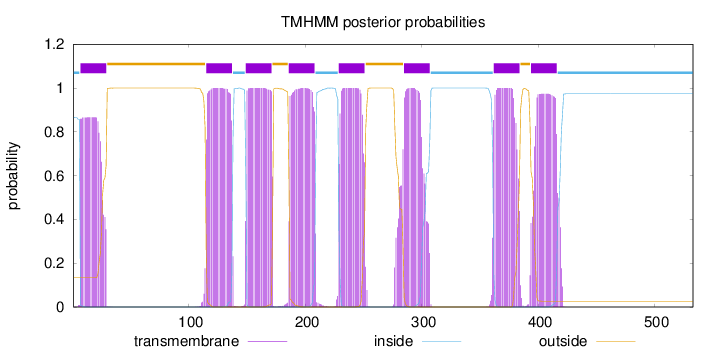
Length:
533
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
174.50603
Exp number, first 60 AAs:
17.72977
Total prob of N-in:
0.86571
POSSIBLE N-term signal
sequence
inside
1 - 6
TMhelix
7 - 29
outside
30 - 114
TMhelix
115 - 137
inside
138 - 148
TMhelix
149 - 171
outside
172 - 185
TMhelix
186 - 208
inside
209 - 228
TMhelix
229 - 251
outside
252 - 284
TMhelix
285 - 307
inside
308 - 361
TMhelix
362 - 384
outside
385 - 393
TMhelix
394 - 416
inside
417 - 533
Population Genetic Test Statistics
Pi
196.491803
Theta
155.222588
Tajima's D
0.513778
CLR
0.301761
CSRT
0.51902404879756
Interpretation
Uncertain
