Pre Gene Modal
BGIBMGA004267
Annotation
PREDICTED:_uncharacterized_protein_LOC106137965_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 4.258
Sequence
CDS
ATGCTGATATTTGATATTTATTTATCAACATTAGTTCGAGTGGTTTTTGTATCATATAATATAATAATGAGTTCAGGATTTATCTCAGAGTCTGAAATTTTGGAATCCAGACGCCGTCGGCAAGAGGAATGGGATAAAGTACGTACAGAAGACCAACCAAGAGAGGCACCCGAAGAGGAGTATGATTCAAGGCCACTGTTTCAACGGCTTGAAGAGCAAAGAATGAAGAAAGATGCTGAATATGAGGAAGCGCATAAACTTAAAAACATGATCCGTGGTTTGGATGATGATGAGGTGGGCTTCCTGGATCTTGTCGAGAGAACAAAAGCTAAAGCAGCTCAGCAAATTTCCATTGAAGAACAGAAGGAAATGCAAGAATTCAGAGAGCGAGTGTCCAATCTAGCTGAGAGTGAAGAAATGACTCGTCTCCGAGCTCAATTGGCTCTGGGTAAAACTCAAACCACAACAACACCGACACAGAAAAACAAACTGCAAGGGTTGATTGTCAGAAAGCGTAAGGCGAGCGAAATAGAAGAGGAAAATCAGAAAGACGCGCCGCCCGCCAAGACCAACGGAGTGGTTTCGAACAATGTTGCAACAAGTAAAATTCAAGCGGATTCCGGGATAGTCGTGGGCGCGTTGAGTGTGGCCGGAGTGTTGCCAGGGTTGGGACACTACCGCTCTGATTCATCTACCGATACAACCGATTCTAGTGACGACGAGAGCGATAAATGTTGCAAGAGGGACTTGCTGGGACGGCGGCCGGAGCACGCGGAGAGCAAGTCGCGCAAAGACGAATCCAAAAGTTAG
Protein
MLIFDIYLSTLVRVVFVSYNIIMSSGFISESEILESRRRRQEEWDKVRTEDQPREAPEEEYDSRPLFQRLEEQRMKKDAEYEEAHKLKNMIRGLDDDEVGFLDLVERTKAKAAQQISIEEQKEMQEFRERVSNLAESEEMTRLRAQLALGKTQTTTTPTQKNKLQGLIVRKRKASEIEEENQKDAPPAKTNGVVSNNVATSKIQADSGIVVGALSVAGVLPGLGHYRSDSSTDTTDSSDDESDKCCKRDLLGRRPEHAESKSRKDESKS
Summary
Uniprot
H9J428
S4P7K2
A0A2H1W6B3
A0A2A4K4F4
A0A0N1IDJ2
A0A2W1BPW7
+ More
A0A0N1PJ77 B0WBG1 B0XKY2 A0A023ELM7 A0A1Q3FDB8 Q16H94 A0A182REK5 A0A182KV89 A0A182M7F1 U5ETC4 A0A182I5T8 A0A182X0J8 A0A182UFV6 A0A182W9L3 A0A182V307 A0A182K3K9 W5JFX7 A0A034WJX1 Q7Q211 A0A182Q603 A0A0A1X1F7 A0A182P9U5 A0A182NV59 E2BZR0 D6WTB2 A0A182S674 A0A182YIW0 A0A084WUH5 W8BKI8 A0A182FVX6 A0A182J8K5 A0A1Y1NBN9 K7J6B9 A0A151INU1 A0A1I8QAA8 N6SSL4 A0A2M3ZAY7 A0A0K8UEX4 A0A2M4BX03 A0A088A8A8 A0A2M4ANF1 A0A2M3ZIN8 A0A2A3EA06 A0A151X8K7 A0A2M3ZAR5 A0A195ELB8 A0A151JSS1 A0A232EJB3 E9IX25 E1ZZX8 A0A2P8YF16 F4W7G5 A0A067R8H1 A0A1L8ECU2 J9JPX6 A0A1L8ECT9 A0A2S2N777 A0A195ATT2 A0A158NPS9 A0A310S790 A0A0M3QUP9 V5I7J8 A0A0J7KTH8 A0A336M7Y0 A0A1L8ECT3 J3JYK4 A0A1I8N1V9 T1PBE9 B4LQ35 B4KM05 A0A1A9WDT6 A0A2J7R827 E0VHB4 V5HK30 A0A1B6FAA3 B4J4Q0 A0A0N8BS45 A0A0P6CJV6 A0A131Y9U3 A0A162RJN6 A0A0P5RCG6 A0A1B6J2L6 A0A0N8ASN1 B3MIH8 B4MQM4 E9HIY3 A0A023FMF9
A0A0N1PJ77 B0WBG1 B0XKY2 A0A023ELM7 A0A1Q3FDB8 Q16H94 A0A182REK5 A0A182KV89 A0A182M7F1 U5ETC4 A0A182I5T8 A0A182X0J8 A0A182UFV6 A0A182W9L3 A0A182V307 A0A182K3K9 W5JFX7 A0A034WJX1 Q7Q211 A0A182Q603 A0A0A1X1F7 A0A182P9U5 A0A182NV59 E2BZR0 D6WTB2 A0A182S674 A0A182YIW0 A0A084WUH5 W8BKI8 A0A182FVX6 A0A182J8K5 A0A1Y1NBN9 K7J6B9 A0A151INU1 A0A1I8QAA8 N6SSL4 A0A2M3ZAY7 A0A0K8UEX4 A0A2M4BX03 A0A088A8A8 A0A2M4ANF1 A0A2M3ZIN8 A0A2A3EA06 A0A151X8K7 A0A2M3ZAR5 A0A195ELB8 A0A151JSS1 A0A232EJB3 E9IX25 E1ZZX8 A0A2P8YF16 F4W7G5 A0A067R8H1 A0A1L8ECU2 J9JPX6 A0A1L8ECT9 A0A2S2N777 A0A195ATT2 A0A158NPS9 A0A310S790 A0A0M3QUP9 V5I7J8 A0A0J7KTH8 A0A336M7Y0 A0A1L8ECT3 J3JYK4 A0A1I8N1V9 T1PBE9 B4LQ35 B4KM05 A0A1A9WDT6 A0A2J7R827 E0VHB4 V5HK30 A0A1B6FAA3 B4J4Q0 A0A0N8BS45 A0A0P6CJV6 A0A131Y9U3 A0A162RJN6 A0A0P5RCG6 A0A1B6J2L6 A0A0N8ASN1 B3MIH8 B4MQM4 E9HIY3 A0A023FMF9
Pubmed
19121390
23622113
26354079
28756777
24945155
17510324
+ More
20966253 20920257 23761445 25348373 12364791 14747013 17210077 25830018 20798317 18362917 19820115 25244985 24438588 24495485 28004739 20075255 23537049 28648823 21282665 29403074 21719571 24845553 21347285 22516182 25315136 17994087 20566863 25765539 21292972
20966253 20920257 23761445 25348373 12364791 14747013 17210077 25830018 20798317 18362917 19820115 25244985 24438588 24495485 28004739 20075255 23537049 28648823 21282665 29403074 21719571 24845553 21347285 22516182 25315136 17994087 20566863 25765539 21292972
EMBL
BABH01041301
GAIX01004559
JAA88001.1
ODYU01006592
SOQ48567.1
NWSH01000141
+ More
PCG79127.1 LADI01012895 KPJ20748.1 KZ149937 PZC77099.1 LADJ01037929 KPJ21325.1 DS231879 EDS42400.1 DS234118 EDS32958.1 GAPW01003451 JAC10147.1 GFDL01009515 JAV25530.1 CH478192 EAT33605.1 EAT33606.1 AXCM01011909 GANO01002838 JAB57033.1 APCN01003502 ADMH02001614 ETN61785.1 GAKP01004320 JAC54632.1 AAAB01008979 EAA13738.2 AXCN02001807 GBXI01009581 JAD04711.1 GL451657 EFN78822.1 KQ971352 EFA06689.1 ATLV01027085 KE525423 KFB53869.1 GAMC01012765 GAMC01012764 JAB93791.1 GEZM01007224 GEZM01007222 JAV95283.1 KQ976914 KYN07133.1 APGK01058319 APGK01058320 KB741288 ENN70654.1 GGFM01004953 MBW25704.1 GDHF01027120 JAI25194.1 GGFJ01008422 MBW57563.1 GGFK01008993 MBW42314.1 GGFM01007686 MBW28437.1 KZ288325 PBC28006.1 KQ982409 KYQ56707.1 GGFM01004797 MBW25548.1 KQ978730 KYN28926.1 KQ982014 KYN30654.1 NNAY01004039 OXU18459.1 GL766616 EFZ14904.1 GL435531 EFN73250.1 PYGN01000648 PSN42858.1 GL887844 EGI69838.1 KK852634 KDR19768.1 GFDG01002343 JAV16456.1 ABLF02036058 GFDG01002344 JAV16455.1 GGMR01000173 MBY12792.1 KQ976745 KYM75462.1 ADTU01022742 KQ766586 OAD53667.1 CP012524 ALC41049.1 GALX01006310 JAB62156.1 LBMM01003354 KMQ93618.1 UFQT01000449 UFQT01003414 SSX24477.1 SSX34978.1 GFDG01002354 JAV16445.1 BT128333 KB632322 AEE63291.1 ERL92358.1 KA646072 AFP60701.1 CH940648 EDW60358.1 CH933808 EDW10794.1 NEVH01006723 PNF36982.1 DS235165 EEB12770.1 GANP01008806 JAB75662.1 GECZ01022642 GECZ01001163 JAS47127.1 JAS68606.1 CH916367 EDW00596.1 GDIQ01150299 JAL01427.1 GDIP01007148 JAM96567.1 GEFM01000531 JAP75265.1 LRGB01000149 KZS20565.1 GDIQ01102712 JAL49014.1 GECU01014298 GECU01007842 JAS93408.1 JAS99864.1 GDIQ01246811 JAK04914.1 CH902619 EDV37026.1 CH963849 EDW74413.1 GL732658 EFX68291.1 GBBK01001580 JAC22902.1
PCG79127.1 LADI01012895 KPJ20748.1 KZ149937 PZC77099.1 LADJ01037929 KPJ21325.1 DS231879 EDS42400.1 DS234118 EDS32958.1 GAPW01003451 JAC10147.1 GFDL01009515 JAV25530.1 CH478192 EAT33605.1 EAT33606.1 AXCM01011909 GANO01002838 JAB57033.1 APCN01003502 ADMH02001614 ETN61785.1 GAKP01004320 JAC54632.1 AAAB01008979 EAA13738.2 AXCN02001807 GBXI01009581 JAD04711.1 GL451657 EFN78822.1 KQ971352 EFA06689.1 ATLV01027085 KE525423 KFB53869.1 GAMC01012765 GAMC01012764 JAB93791.1 GEZM01007224 GEZM01007222 JAV95283.1 KQ976914 KYN07133.1 APGK01058319 APGK01058320 KB741288 ENN70654.1 GGFM01004953 MBW25704.1 GDHF01027120 JAI25194.1 GGFJ01008422 MBW57563.1 GGFK01008993 MBW42314.1 GGFM01007686 MBW28437.1 KZ288325 PBC28006.1 KQ982409 KYQ56707.1 GGFM01004797 MBW25548.1 KQ978730 KYN28926.1 KQ982014 KYN30654.1 NNAY01004039 OXU18459.1 GL766616 EFZ14904.1 GL435531 EFN73250.1 PYGN01000648 PSN42858.1 GL887844 EGI69838.1 KK852634 KDR19768.1 GFDG01002343 JAV16456.1 ABLF02036058 GFDG01002344 JAV16455.1 GGMR01000173 MBY12792.1 KQ976745 KYM75462.1 ADTU01022742 KQ766586 OAD53667.1 CP012524 ALC41049.1 GALX01006310 JAB62156.1 LBMM01003354 KMQ93618.1 UFQT01000449 UFQT01003414 SSX24477.1 SSX34978.1 GFDG01002354 JAV16445.1 BT128333 KB632322 AEE63291.1 ERL92358.1 KA646072 AFP60701.1 CH940648 EDW60358.1 CH933808 EDW10794.1 NEVH01006723 PNF36982.1 DS235165 EEB12770.1 GANP01008806 JAB75662.1 GECZ01022642 GECZ01001163 JAS47127.1 JAS68606.1 CH916367 EDW00596.1 GDIQ01150299 JAL01427.1 GDIP01007148 JAM96567.1 GEFM01000531 JAP75265.1 LRGB01000149 KZS20565.1 GDIQ01102712 JAL49014.1 GECU01014298 GECU01007842 JAS93408.1 JAS99864.1 GDIQ01246811 JAK04914.1 CH902619 EDV37026.1 CH963849 EDW74413.1 GL732658 EFX68291.1 GBBK01001580 JAC22902.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000002320
UP000008820
+ More
UP000075900 UP000075882 UP000075883 UP000075840 UP000076407 UP000075902 UP000075920 UP000075903 UP000075881 UP000000673 UP000007062 UP000075886 UP000075885 UP000075884 UP000008237 UP000007266 UP000075901 UP000076408 UP000030765 UP000069272 UP000075880 UP000002358 UP000078542 UP000095300 UP000019118 UP000005203 UP000242457 UP000075809 UP000078492 UP000078541 UP000215335 UP000000311 UP000245037 UP000007755 UP000027135 UP000007819 UP000078540 UP000005205 UP000092553 UP000036403 UP000030742 UP000095301 UP000008792 UP000009192 UP000091820 UP000235965 UP000009046 UP000001070 UP000076858 UP000007801 UP000007798 UP000000305
UP000075900 UP000075882 UP000075883 UP000075840 UP000076407 UP000075902 UP000075920 UP000075903 UP000075881 UP000000673 UP000007062 UP000075886 UP000075885 UP000075884 UP000008237 UP000007266 UP000075901 UP000076408 UP000030765 UP000069272 UP000075880 UP000002358 UP000078542 UP000095300 UP000019118 UP000005203 UP000242457 UP000075809 UP000078492 UP000078541 UP000215335 UP000000311 UP000245037 UP000007755 UP000027135 UP000007819 UP000078540 UP000005205 UP000092553 UP000036403 UP000030742 UP000095301 UP000008792 UP000009192 UP000091820 UP000235965 UP000009046 UP000001070 UP000076858 UP000007801 UP000007798 UP000000305
PRIDE
ProteinModelPortal
H9J428
S4P7K2
A0A2H1W6B3
A0A2A4K4F4
A0A0N1IDJ2
A0A2W1BPW7
+ More
A0A0N1PJ77 B0WBG1 B0XKY2 A0A023ELM7 A0A1Q3FDB8 Q16H94 A0A182REK5 A0A182KV89 A0A182M7F1 U5ETC4 A0A182I5T8 A0A182X0J8 A0A182UFV6 A0A182W9L3 A0A182V307 A0A182K3K9 W5JFX7 A0A034WJX1 Q7Q211 A0A182Q603 A0A0A1X1F7 A0A182P9U5 A0A182NV59 E2BZR0 D6WTB2 A0A182S674 A0A182YIW0 A0A084WUH5 W8BKI8 A0A182FVX6 A0A182J8K5 A0A1Y1NBN9 K7J6B9 A0A151INU1 A0A1I8QAA8 N6SSL4 A0A2M3ZAY7 A0A0K8UEX4 A0A2M4BX03 A0A088A8A8 A0A2M4ANF1 A0A2M3ZIN8 A0A2A3EA06 A0A151X8K7 A0A2M3ZAR5 A0A195ELB8 A0A151JSS1 A0A232EJB3 E9IX25 E1ZZX8 A0A2P8YF16 F4W7G5 A0A067R8H1 A0A1L8ECU2 J9JPX6 A0A1L8ECT9 A0A2S2N777 A0A195ATT2 A0A158NPS9 A0A310S790 A0A0M3QUP9 V5I7J8 A0A0J7KTH8 A0A336M7Y0 A0A1L8ECT3 J3JYK4 A0A1I8N1V9 T1PBE9 B4LQ35 B4KM05 A0A1A9WDT6 A0A2J7R827 E0VHB4 V5HK30 A0A1B6FAA3 B4J4Q0 A0A0N8BS45 A0A0P6CJV6 A0A131Y9U3 A0A162RJN6 A0A0P5RCG6 A0A1B6J2L6 A0A0N8ASN1 B3MIH8 B4MQM4 E9HIY3 A0A023FMF9
A0A0N1PJ77 B0WBG1 B0XKY2 A0A023ELM7 A0A1Q3FDB8 Q16H94 A0A182REK5 A0A182KV89 A0A182M7F1 U5ETC4 A0A182I5T8 A0A182X0J8 A0A182UFV6 A0A182W9L3 A0A182V307 A0A182K3K9 W5JFX7 A0A034WJX1 Q7Q211 A0A182Q603 A0A0A1X1F7 A0A182P9U5 A0A182NV59 E2BZR0 D6WTB2 A0A182S674 A0A182YIW0 A0A084WUH5 W8BKI8 A0A182FVX6 A0A182J8K5 A0A1Y1NBN9 K7J6B9 A0A151INU1 A0A1I8QAA8 N6SSL4 A0A2M3ZAY7 A0A0K8UEX4 A0A2M4BX03 A0A088A8A8 A0A2M4ANF1 A0A2M3ZIN8 A0A2A3EA06 A0A151X8K7 A0A2M3ZAR5 A0A195ELB8 A0A151JSS1 A0A232EJB3 E9IX25 E1ZZX8 A0A2P8YF16 F4W7G5 A0A067R8H1 A0A1L8ECU2 J9JPX6 A0A1L8ECT9 A0A2S2N777 A0A195ATT2 A0A158NPS9 A0A310S790 A0A0M3QUP9 V5I7J8 A0A0J7KTH8 A0A336M7Y0 A0A1L8ECT3 J3JYK4 A0A1I8N1V9 T1PBE9 B4LQ35 B4KM05 A0A1A9WDT6 A0A2J7R827 E0VHB4 V5HK30 A0A1B6FAA3 B4J4Q0 A0A0N8BS45 A0A0P6CJV6 A0A131Y9U3 A0A162RJN6 A0A0P5RCG6 A0A1B6J2L6 A0A0N8ASN1 B3MIH8 B4MQM4 E9HIY3 A0A023FMF9
Ontologies
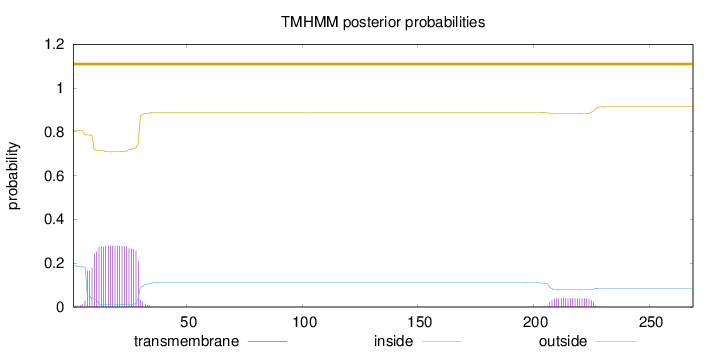
Topology
Length:
269
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.80017
Exp number, first 60 AAs:
6.03401
Total prob of N-in:
0.19250
outside
1 - 269
Population Genetic Test Statistics
Pi
252.228393
Theta
71.552793
Tajima's D
0.758097
CLR
0.003157
CSRT
0.592020398980051
Interpretation
Uncertain
