Gene
KWMTBOMO11834
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.734
Sequence
CDS
ATGACTACTCTGGAGATCTCGTTGGTCGGGAGTGTGGTTAACATTGGAGCATTGGTAGCCACGCCTTTCTGTACTTACTTCTTAAATAAGTTGGGGCGGAAATACACATGTATGCTCTTTGGCGTGCCTTATGCGATGTCATGGGCGATACTGTCAGTATCTAATTATGTTCCTTTTGTCATCGTCGCTTTGGCTTTGAGTGGAATTGGCGCTGCGGGTGTACCGGCCTCAACAATTTATATCTCGGAGATATCAGATGATTCAATCAGAGGAATTTTAACGTCAAATGCGATATCAGGATATTTTCTCGGACTTTTTCTCTCCTACTCCTGGGGAGGATACTTGAGCTATTACAACGTAGTCCACTTGCACTTGGGTCTATGCGTGCTGTATATTATAATGGTTGGTTCTTTGAAGGATTCCCCGGTTTTCTTGATGCAAACCAACAAAGAAAAGGAGGCGGCGAAATCGATTGCCTTTTATCGGTGTGTGGACGTCACATCTAAAGAAGTGGAGATAGAACTAGCCAATATCCGCCTGCAGCTTGATCCTCGTCGAGAGAATATTCTAGAAGGGAAAAATGATATGACAGTCACAAAAGAATTGTTAAAGATTAAATCAGAACCAGCCGAAGCGAAATCAGAATGGAAATTTTTAATCAATTCCCCGTCATCAATTCGAGCTTTGACTATCGCTATTATCACGATGACGTACACGGTGCTCATGGGAGTGGTGGTGCTCCAGGTATACGCCGAACCACTATTCAGAGAAGCAGTCCCATCGATGGAGCCGAACCAGTTGACGATTTACCTGGCGATAGATTTCATTGTGGGCAGTCTTATTTGTGGAGTACTCATTGACAGATTGGGGCGTAAGTACCTGCTGATCATCACCACAGTCCTGACCAGCATATGTATATTACTGATGGCGACTCAGCTGCAGTTCAGCTGGGCTCCACAGTGGACGACAGCGGGCTTCATGTACGCCTTCTCCTTCTTCTACAACTTGGGACCGGCGACGATTCCGTTCGTGATCGCTGCCGAGTTCTTTGTGCCAGAAGTCCGAGGTCTGTGCAATTGTTTGATCAACGCCTGTACTTGGCTCATGAATTTTGTGACGCTTCAATTATTCTCACAACTCGTCGATTGGGTCGGCCTGGCTCCTCTCTTCTACCTCTTCAGTTTCTTCGGATTCCTTGGAGCGGTCTTTTGTCAATTTTACTTGCCGGAGACTAAGGGTATGCCCATCGAAGCCATTCCGTTGCTCTTCATTAAGAAAGATAAGAGAAGACATTATAATATCTAA
Protein
MTTLEISLVGSVVNIGALVATPFCTYFLNKLGRKYTCMLFGVPYAMSWAILSVSNYVPFVIVALALSGIGAAGVPASTIYISEISDDSIRGILTSNAISGYFLGLFLSYSWGGYLSYYNVVHLHLGLCVLYIIMVGSLKDSPVFLMQTNKEKEAAKSIAFYRCVDVTSKEVEIELANIRLQLDPRRENILEGKNDMTVTKELLKIKSEPAEAKSEWKFLINSPSSIRALTIAIITMTYTVLMGVVVLQVYAEPLFREAVPSMEPNQLTIYLAIDFIVGSLICGVLIDRLGRKYLLIITTVLTSICILLMATQLQFSWAPQWTTAGFMYAFSFFYNLGPATIPFVIAAEFFVPEVRGLCNCLINACTWLMNFVTLQLFSQLVDWVGLAPLFYLFSFFGFLGAVFCQFYLPETKGMPIEAIPLLFIKKDKRRHYNI
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
A0A2A4JUG4
A0A0N1PKK0
A0A0N1IQK5
A0A2W1BZ84
A0A2H1WYH9
A0A212F787
+ More
A0A2W1BPY0 A0A2A4JV69 H9JWY9 A0A3S2M6I8 A0A2W1BTX0 A0A2A4JW98 A0A194Q6Z4 A0A0N0PEG4 A0A212F769 A0A194QBG3 A0A194Q5E1 A0A2H1VA59 A0A194Q5E7 H9JVU1 A0A2A4JYQ5 A0A2W1BWR1 A0A3S2LEN2 A0A0L7KPP9 A0A3S2LR68 A0A212F6E9 A0A2W1BV94 A0A0N0PED5 A0A2A4JW00 A0A0N0PF14 A0A2H1X1H2 A0A2A4K0E5 A0A2W1BTZ9 H9JWZ0 A0A0N1PIZ3 A0A2H1WVK7 A0A1B6GYS0 A0A1B6KIF4 A0A0L7L2W9 A0A2J7PYX6 A0A2J7R8L3 A0A336MWL1 A0A067R867 A0A336MDM9 A0A1S4FFK9 Q173J5 A0A0A1WNR4 A0A1B6FS48 A0A0C9Q8K4 A0A2P8Z801 A0A182TXA5 B0WPK5 A0A182JZA6 A0A182I799 A0A1B6CY32 A0A1B6DME3 B0WPK6 A0A1Q3F5A2 Q7PWP0 A0A1S4GZI9 W8BY63 A0A2M4A7E4 A0A182LMV4 A0A2M4A741 A0A2M4BLL7 A0A182XNP0 A0A2J7R8M9 A0A182UZ66 A0A182P8A2 A0A2M4BLK9 A0A2M4A739 A0A0M4EUF1 A0A2M3Z428 A0A2M3Z412 A0A2M3ZH26 A0A026WMM0 A0A084W7N7 A0A182NGP0 A0A182QRF9 A0A1L8DIV6 A0A182NAM9 A0A0A8J8K5 A0A182F539 W5JMX1 E9J4B0 A0A2M4CQC1 A0A1Q3FS38 A0A2M4CQF1 A0A182MPL4 A0A226EKQ9 A0A182UH29 A0A182R408 A0A3L8D924 A0A2P8ZLZ6 A0A0C9RQR2 A0A1B6JA16
A0A2W1BPY0 A0A2A4JV69 H9JWY9 A0A3S2M6I8 A0A2W1BTX0 A0A2A4JW98 A0A194Q6Z4 A0A0N0PEG4 A0A212F769 A0A194QBG3 A0A194Q5E1 A0A2H1VA59 A0A194Q5E7 H9JVU1 A0A2A4JYQ5 A0A2W1BWR1 A0A3S2LEN2 A0A0L7KPP9 A0A3S2LR68 A0A212F6E9 A0A2W1BV94 A0A0N0PED5 A0A2A4JW00 A0A0N0PF14 A0A2H1X1H2 A0A2A4K0E5 A0A2W1BTZ9 H9JWZ0 A0A0N1PIZ3 A0A2H1WVK7 A0A1B6GYS0 A0A1B6KIF4 A0A0L7L2W9 A0A2J7PYX6 A0A2J7R8L3 A0A336MWL1 A0A067R867 A0A336MDM9 A0A1S4FFK9 Q173J5 A0A0A1WNR4 A0A1B6FS48 A0A0C9Q8K4 A0A2P8Z801 A0A182TXA5 B0WPK5 A0A182JZA6 A0A182I799 A0A1B6CY32 A0A1B6DME3 B0WPK6 A0A1Q3F5A2 Q7PWP0 A0A1S4GZI9 W8BY63 A0A2M4A7E4 A0A182LMV4 A0A2M4A741 A0A2M4BLL7 A0A182XNP0 A0A2J7R8M9 A0A182UZ66 A0A182P8A2 A0A2M4BLK9 A0A2M4A739 A0A0M4EUF1 A0A2M3Z428 A0A2M3Z412 A0A2M3ZH26 A0A026WMM0 A0A084W7N7 A0A182NGP0 A0A182QRF9 A0A1L8DIV6 A0A182NAM9 A0A0A8J8K5 A0A182F539 W5JMX1 E9J4B0 A0A2M4CQC1 A0A1Q3FS38 A0A2M4CQF1 A0A182MPL4 A0A226EKQ9 A0A182UH29 A0A182R408 A0A3L8D924 A0A2P8ZLZ6 A0A0C9RQR2 A0A1B6JA16
Pubmed
EMBL
NWSH01000558
PCG75665.1
LADJ01037929
KPJ21328.1
LADI01012895
KPJ20745.1
+ More
KZ149937 PZC77103.1 ODYU01011969 SOQ58016.1 AGBW02009924 OWR49569.1 PZC77102.1 PCG75666.1 BABH01043549 RSAL01000025 RVE52104.1 PZC77105.1 PCG75662.1 KQ459460 KPJ00770.1 KQ459838 KPJ19748.1 OWR49568.1 KPJ00771.1 KPJ00767.1 ODYU01001454 SOQ37676.1 KPJ00768.1 BABH01040660 NWSH01000350 PCG77147.1 PZC77116.1 RVE52115.1 JTDY01007385 KOB65252.1 RVE52106.1 AGBW02010035 OWR49315.1 PZC77117.1 KPJ19751.1 PCG75663.1 KPJ20744.1 ODYU01012723 SOQ59169.1 PCG77150.1 PZC77104.1 BABH01041799 BABH01041800 BABH01041801 KQ460009 KPJ18482.1 ODYU01011381 SOQ57088.1 GECZ01003576 GECZ01002212 JAS66193.1 JAS67557.1 GEBQ01028740 JAT11237.1 JTDY01003373 KOB69656.1 NEVH01020342 PNF21526.1 NEVH01006721 PNF37174.1 UFQT01003140 SSX34636.1 KK852870 KDR14605.1 UFQT01000924 SSX28030.1 CH477423 EAT41201.1 GBXI01014229 JAD00063.1 GECZ01016734 JAS53035.1 GBYB01010573 JAG80340.1 PYGN01000154 PSN52620.1 DS232026 EDS32406.1 APCN01003374 GEDC01018964 JAS18334.1 GEDC01010470 JAS26828.1 EDS32407.1 GFDL01012362 JAV22683.1 AAAB01008984 EAA14882.4 GAMC01012366 JAB94189.1 GGFK01003378 MBW36699.1 GGFK01003296 MBW36617.1 GGFJ01004829 MBW53970.1 PNF37175.1 GGFJ01004828 MBW53969.1 GGFK01003278 MBW36599.1 CP012524 ALC41045.1 GGFM01002530 MBW23281.1 GGFM01002508 MBW23259.1 GGFM01007009 MBW27760.1 KK107151 QOIP01000003 EZA57322.1 RLU25205.1 ATLV01021268 KE525315 KFB46231.1 AXCN02002080 GFDF01007794 JAV06290.1 AB901068 BAQ02368.1 ADMH02001096 ETN64119.1 GL768090 EFZ12343.1 GGFL01003356 MBW67534.1 GFDL01004625 JAV30420.1 GGFL01003355 MBW67533.1 AXCM01000345 LNIX01000003 OXA58039.1 QOIP01000011 RLU16997.1 PYGN01000019 PSN57527.1 GBYB01009586 JAG79353.1 GECU01011708 JAS95998.1
KZ149937 PZC77103.1 ODYU01011969 SOQ58016.1 AGBW02009924 OWR49569.1 PZC77102.1 PCG75666.1 BABH01043549 RSAL01000025 RVE52104.1 PZC77105.1 PCG75662.1 KQ459460 KPJ00770.1 KQ459838 KPJ19748.1 OWR49568.1 KPJ00771.1 KPJ00767.1 ODYU01001454 SOQ37676.1 KPJ00768.1 BABH01040660 NWSH01000350 PCG77147.1 PZC77116.1 RVE52115.1 JTDY01007385 KOB65252.1 RVE52106.1 AGBW02010035 OWR49315.1 PZC77117.1 KPJ19751.1 PCG75663.1 KPJ20744.1 ODYU01012723 SOQ59169.1 PCG77150.1 PZC77104.1 BABH01041799 BABH01041800 BABH01041801 KQ460009 KPJ18482.1 ODYU01011381 SOQ57088.1 GECZ01003576 GECZ01002212 JAS66193.1 JAS67557.1 GEBQ01028740 JAT11237.1 JTDY01003373 KOB69656.1 NEVH01020342 PNF21526.1 NEVH01006721 PNF37174.1 UFQT01003140 SSX34636.1 KK852870 KDR14605.1 UFQT01000924 SSX28030.1 CH477423 EAT41201.1 GBXI01014229 JAD00063.1 GECZ01016734 JAS53035.1 GBYB01010573 JAG80340.1 PYGN01000154 PSN52620.1 DS232026 EDS32406.1 APCN01003374 GEDC01018964 JAS18334.1 GEDC01010470 JAS26828.1 EDS32407.1 GFDL01012362 JAV22683.1 AAAB01008984 EAA14882.4 GAMC01012366 JAB94189.1 GGFK01003378 MBW36699.1 GGFK01003296 MBW36617.1 GGFJ01004829 MBW53970.1 PNF37175.1 GGFJ01004828 MBW53969.1 GGFK01003278 MBW36599.1 CP012524 ALC41045.1 GGFM01002530 MBW23281.1 GGFM01002508 MBW23259.1 GGFM01007009 MBW27760.1 KK107151 QOIP01000003 EZA57322.1 RLU25205.1 ATLV01021268 KE525315 KFB46231.1 AXCN02002080 GFDF01007794 JAV06290.1 AB901068 BAQ02368.1 ADMH02001096 ETN64119.1 GL768090 EFZ12343.1 GGFL01003356 MBW67534.1 GFDL01004625 JAV30420.1 GGFL01003355 MBW67533.1 AXCM01000345 LNIX01000003 OXA58039.1 QOIP01000011 RLU16997.1 PYGN01000019 PSN57527.1 GBYB01009586 JAG79353.1 GECU01011708 JAS95998.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000005204
UP000283053
+ More
UP000037510 UP000235965 UP000027135 UP000008820 UP000245037 UP000075902 UP000002320 UP000075881 UP000075840 UP000007062 UP000075882 UP000076407 UP000075903 UP000075885 UP000092553 UP000053097 UP000279307 UP000030765 UP000075884 UP000075886 UP000069272 UP000000673 UP000075883 UP000198287 UP000075900
UP000037510 UP000235965 UP000027135 UP000008820 UP000245037 UP000075902 UP000002320 UP000075881 UP000075840 UP000007062 UP000075882 UP000076407 UP000075903 UP000075885 UP000092553 UP000053097 UP000279307 UP000030765 UP000075884 UP000075886 UP000069272 UP000000673 UP000075883 UP000198287 UP000075900
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2A4JUG4
A0A0N1PKK0
A0A0N1IQK5
A0A2W1BZ84
A0A2H1WYH9
A0A212F787
+ More
A0A2W1BPY0 A0A2A4JV69 H9JWY9 A0A3S2M6I8 A0A2W1BTX0 A0A2A4JW98 A0A194Q6Z4 A0A0N0PEG4 A0A212F769 A0A194QBG3 A0A194Q5E1 A0A2H1VA59 A0A194Q5E7 H9JVU1 A0A2A4JYQ5 A0A2W1BWR1 A0A3S2LEN2 A0A0L7KPP9 A0A3S2LR68 A0A212F6E9 A0A2W1BV94 A0A0N0PED5 A0A2A4JW00 A0A0N0PF14 A0A2H1X1H2 A0A2A4K0E5 A0A2W1BTZ9 H9JWZ0 A0A0N1PIZ3 A0A2H1WVK7 A0A1B6GYS0 A0A1B6KIF4 A0A0L7L2W9 A0A2J7PYX6 A0A2J7R8L3 A0A336MWL1 A0A067R867 A0A336MDM9 A0A1S4FFK9 Q173J5 A0A0A1WNR4 A0A1B6FS48 A0A0C9Q8K4 A0A2P8Z801 A0A182TXA5 B0WPK5 A0A182JZA6 A0A182I799 A0A1B6CY32 A0A1B6DME3 B0WPK6 A0A1Q3F5A2 Q7PWP0 A0A1S4GZI9 W8BY63 A0A2M4A7E4 A0A182LMV4 A0A2M4A741 A0A2M4BLL7 A0A182XNP0 A0A2J7R8M9 A0A182UZ66 A0A182P8A2 A0A2M4BLK9 A0A2M4A739 A0A0M4EUF1 A0A2M3Z428 A0A2M3Z412 A0A2M3ZH26 A0A026WMM0 A0A084W7N7 A0A182NGP0 A0A182QRF9 A0A1L8DIV6 A0A182NAM9 A0A0A8J8K5 A0A182F539 W5JMX1 E9J4B0 A0A2M4CQC1 A0A1Q3FS38 A0A2M4CQF1 A0A182MPL4 A0A226EKQ9 A0A182UH29 A0A182R408 A0A3L8D924 A0A2P8ZLZ6 A0A0C9RQR2 A0A1B6JA16
A0A2W1BPY0 A0A2A4JV69 H9JWY9 A0A3S2M6I8 A0A2W1BTX0 A0A2A4JW98 A0A194Q6Z4 A0A0N0PEG4 A0A212F769 A0A194QBG3 A0A194Q5E1 A0A2H1VA59 A0A194Q5E7 H9JVU1 A0A2A4JYQ5 A0A2W1BWR1 A0A3S2LEN2 A0A0L7KPP9 A0A3S2LR68 A0A212F6E9 A0A2W1BV94 A0A0N0PED5 A0A2A4JW00 A0A0N0PF14 A0A2H1X1H2 A0A2A4K0E5 A0A2W1BTZ9 H9JWZ0 A0A0N1PIZ3 A0A2H1WVK7 A0A1B6GYS0 A0A1B6KIF4 A0A0L7L2W9 A0A2J7PYX6 A0A2J7R8L3 A0A336MWL1 A0A067R867 A0A336MDM9 A0A1S4FFK9 Q173J5 A0A0A1WNR4 A0A1B6FS48 A0A0C9Q8K4 A0A2P8Z801 A0A182TXA5 B0WPK5 A0A182JZA6 A0A182I799 A0A1B6CY32 A0A1B6DME3 B0WPK6 A0A1Q3F5A2 Q7PWP0 A0A1S4GZI9 W8BY63 A0A2M4A7E4 A0A182LMV4 A0A2M4A741 A0A2M4BLL7 A0A182XNP0 A0A2J7R8M9 A0A182UZ66 A0A182P8A2 A0A2M4BLK9 A0A2M4A739 A0A0M4EUF1 A0A2M3Z428 A0A2M3Z412 A0A2M3ZH26 A0A026WMM0 A0A084W7N7 A0A182NGP0 A0A182QRF9 A0A1L8DIV6 A0A182NAM9 A0A0A8J8K5 A0A182F539 W5JMX1 E9J4B0 A0A2M4CQC1 A0A1Q3FS38 A0A2M4CQF1 A0A182MPL4 A0A226EKQ9 A0A182UH29 A0A182R408 A0A3L8D924 A0A2P8ZLZ6 A0A0C9RQR2 A0A1B6JA16
PDB
5EQI
E-value=8.25489e-11,
Score=162
Ontologies
GO
PANTHER
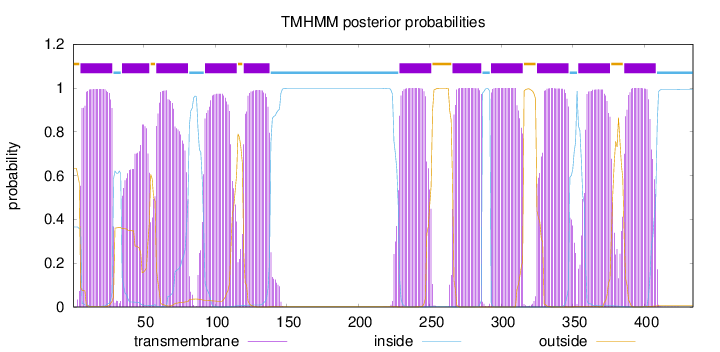
Topology
Length:
434
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
230.29567
Exp number, first 60 AAs:
39.60536
Total prob of N-in:
0.36577
POSSIBLE N-term signal
sequence
outside
1 - 5
TMhelix
6 - 28
inside
29 - 34
TMhelix
35 - 54
outside
55 - 58
TMhelix
59 - 81
inside
82 - 92
TMhelix
93 - 115
outside
116 - 119
TMhelix
120 - 138
inside
139 - 228
TMhelix
229 - 251
outside
252 - 265
TMhelix
266 - 286
inside
287 - 292
TMhelix
293 - 315
outside
316 - 324
TMhelix
325 - 347
inside
348 - 353
TMhelix
354 - 376
outside
377 - 385
TMhelix
386 - 408
inside
409 - 434
Population Genetic Test Statistics
Pi
187.359827
Theta
158.360149
Tajima's D
0.163904
CLR
0.123412
CSRT
0.413829308534573
Interpretation
Uncertain
