Gene
KWMTBOMO11831
Pre Gene Modal
BGIBMGA014449
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.89
Sequence
CDS
ATGATACGTTGCCAGATACTATATATATTTAAATTATTAACGGAATTATTTTACAGGAACCGCATCGAGAGAACCGCTTCAAAACAATGGCGTGGCTCGGTGCGGCGCATCATCTCCGCAACTGTCTATAACCTGTCCTGTTTCACACACGGGTGTAGCACCGGCTGGGTGTCCGGAGTCTTAGGAAACGAAGCGTTGACGGGCGGGGCGTGGCTGGCCGCCCTGCCGTGCCTGGTAGCACTACCGGCAGCTCCATTCTTCGCCGTGCTGGCTGACGCGAGGGGAAGGAAGGCGGGCGCTTTTTGCATTTGCATCAGTTTTATCTTGAGCTGGTCTTTAGCGGCGTGGTGTGGTCCGCGAGGAGTATGGGCTGCCAGGGTTGCAGCCGGCGCAGGGGGTGCGGGCGCGCTTGCCCTCGCACCCCTGTACTGCGCGGAGATAGCGCCACGCACGAGAGGGCTGGCTGCGATGCCAGCACTGGCTTGCAGTTGCGGCATCCTGTTTGCTTACGCGGCAGGTGGCATGCTATCAGCGCACGCATTGTCCTTATCAATGGCCGTACCGCCTTCCGCACTCCTGGTGTCGCTGATATGGCTGCCAGAGACACCTTCGTTTCTGATCAGCATCGGAAGGGTTCAGGATGCCGCAAAGATAATTTGCTGGTTCGACGGCTCCGATTTCAGGGAAGATCTCACCGATGTCATAGAACAACAAGAGGTGAGGATCAGGACCTCAGACTGCTACGGGAGTAGACGGGAGGCGATGCGACGTCAAGACTCTGACGCGTTCCAGCCCATGTTGAAGCGTAGCAGCGGCTTAGAATCTAACAGCGATAAAAAAACCGAGCAATCAGCTTGCAAAGAGTTATTTAGGCATCGCCGCTCCCGGCGAGCTCTCCTCTCGTGTGCGGTGCTCATCTGCGCGGCAGCCGGATCTGGTGCAGGGGCTGTCAACAGCTTCGCAGCGGCAGTACTGAGGCACTCCTCACACGCTGTGCCGCTTCTGAACATGACAGCTTACAACTTCACGATCAACGATGGGTCTCTTCCAAGGCCACTATTCGAGACCTCGGAAGCTGGTGCAATACTTTGTGGTGCCGCATTGGTACTTGGAGCTACGATAGCTACGATTACTGTTGACAAACTTGGAAGGAAAATGCTCCTACTGTGGTCCTGCTCAGGGATAACGTGCTGCCTGATCGTTCTCGGGGTGTACTGCGACCCCCACCTGCGCCTGCACTCCTGGTACTCGCAGCGACTGTGGCCTTACACAGACGAAGACACCCAATCAAAGGAATTGATTGAAGCTTTTAATATTACGAGTAACGTAAACTTCACAAAGCCCTGGGATTATAGGATTAGTGTGGAAAGTGAGAATTTGACTGACAGCTTCATGAGGAGTACGAAGGCTGCTCGGACACATGCAGAGGAGCAAAGGATGTGGGCCCCCGTGGCTTTGCTATCTACAGTATTATTTTTGTACAACATTGGCCTGGGTTCCGTGCCTTTTGTACTCGTCTCGGAGCTTTTTTCGATAAATGTCCGTAGTCTGGCATCAAGTCTGCTCATTTCATGGACGTGGCTGAGCAGCTTTCTCCTTCTGAGGTACTACGGTACTATTGCTGTCTCTCTCGGTCTCCACGGAACCTATTACATATCAGCTTCCATCACGTTTCTGGCATCCTTGTACATATTCCTGGTCTTGCCCGAAACGAAAGGGAAATCGCAAGAGCAGATTGAAGAAATTCTCGACGGGCCAATGTTGGTTTTGAGGAGCACGAAGAAGAACCAAAACCAGCGATGA
Protein
MIRCQILYIFKLLTELFYRNRIERTASKQWRGSVRRIISATVYNLSCFTHGCSTGWVSGVLGNEALTGGAWLAALPCLVALPAAPFFAVLADARGRKAGAFCICISFILSWSLAAWCGPRGVWAARVAAGAGGAGALALAPLYCAEIAPRTRGLAAMPALACSCGILFAYAAGGMLSAHALSLSMAVPPSALLVSLIWLPETPSFLISIGRVQDAAKIICWFDGSDFREDLTDVIEQQEVRIRTSDCYGSRREAMRRQDSDAFQPMLKRSSGLESNSDKKTEQSACKELFRHRRSRRALLSCAVLICAAAGSGAGAVNSFAAAVLRHSSHAVPLLNMTAYNFTINDGSLPRPLFETSEAGAILCGAALVLGATIATITVDKLGRKMLLLWSCSGITCCLIVLGVYCDPHLRLHSWYSQRLWPYTDEDTQSKELIEAFNITSNVNFTKPWDYRISVESENLTDSFMRSTKAARTHAEEQRMWAPVALLSTVLFLYNIGLGSVPFVLVSELFSINVRSLASSLLISWTWLSSFLLLRYYGTIAVSLGLHGTYYISASITFLASLYIFLVLPETKGKSQEQIEEILDGPMLVLRSTKKNQNQR
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9JY31
A0A2H1V6U2
A0A2A4JCE7
A0A0N1PHI9
A0A0L7KA11
A0A2P8ZM01
+ More
A0A026WMM0 E2BT94 F4WBY5 V5GV83 A0A195BM51 A0A158NZ11 A0A026WP92 A0A3L8DXN6 A0A195FTD3 A0A3Q3E8J9 A0A3Q3EBD5 A0A151J628 A0A226EFB2 A0A151IEK9 A0A195FPC4 E0VNX8 A0A3Q3JG04 A0A3P8W6P3 A0A2G5BAM0 A0A3Q2C7A2 A0A1Y1MRS8 E2A9Z9 A0A084VRC5 A0A182MI44 Q7PR34 A0A182LFS7 D6X033 A0A3Q3KU25 A0A182N634
A0A026WMM0 E2BT94 F4WBY5 V5GV83 A0A195BM51 A0A158NZ11 A0A026WP92 A0A3L8DXN6 A0A195FTD3 A0A3Q3E8J9 A0A3Q3EBD5 A0A151J628 A0A226EFB2 A0A151IEK9 A0A195FPC4 E0VNX8 A0A3Q3JG04 A0A3P8W6P3 A0A2G5BAM0 A0A3Q2C7A2 A0A1Y1MRS8 E2A9Z9 A0A084VRC5 A0A182MI44 Q7PR34 A0A182LFS7 D6X033 A0A3Q3KU25 A0A182N634
Pubmed
EMBL
BABH01041989
BABH01041990
ODYU01000976
SOQ36541.1
NWSH01001935
PCG69631.1
+ More
KQ459838 KPJ19746.1 JTDY01010193 KOB60233.1 PYGN01000019 PSN57531.1 KK107151 QOIP01000003 EZA57322.1 RLU25205.1 GL450384 EFN81096.1 GL888067 EGI68274.1 GALX01004323 JAB64143.1 KQ976444 KYM86198.1 ADTU01004466 KK107139 EZA57783.1 RLU25240.1 KQ981276 KYN43567.1 KQ979883 KYN18677.1 LNIX01000004 OXA56232.1 KQ977889 KYM99007.1 KQ981382 KYN42246.1 DS235354 EEB15084.1 KZ303502 PIA16050.1 GEZM01029704 JAV85987.1 GL437987 EFN69767.1 ATLV01015604 KE525023 KFB40519.1 AXCM01000884 AAAB01008859 EAA07901.5 KQ971364 EFA10054.1
KQ459838 KPJ19746.1 JTDY01010193 KOB60233.1 PYGN01000019 PSN57531.1 KK107151 QOIP01000003 EZA57322.1 RLU25205.1 GL450384 EFN81096.1 GL888067 EGI68274.1 GALX01004323 JAB64143.1 KQ976444 KYM86198.1 ADTU01004466 KK107139 EZA57783.1 RLU25240.1 KQ981276 KYN43567.1 KQ979883 KYN18677.1 LNIX01000004 OXA56232.1 KQ977889 KYM99007.1 KQ981382 KYN42246.1 DS235354 EEB15084.1 KZ303502 PIA16050.1 GEZM01029704 JAV85987.1 GL437987 EFN69767.1 ATLV01015604 KE525023 KFB40519.1 AXCM01000884 AAAB01008859 EAA07901.5 KQ971364 EFA10054.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000037510
UP000245037
UP000053097
+ More
UP000279307 UP000008237 UP000007755 UP000078540 UP000005205 UP000078541 UP000261660 UP000078492 UP000198287 UP000078542 UP000009046 UP000261600 UP000265120 UP000242474 UP000265020 UP000000311 UP000030765 UP000075883 UP000007062 UP000075882 UP000007266 UP000261640 UP000075884
UP000279307 UP000008237 UP000007755 UP000078540 UP000005205 UP000078541 UP000261660 UP000078492 UP000198287 UP000078542 UP000009046 UP000261600 UP000265120 UP000242474 UP000265020 UP000000311 UP000030765 UP000075883 UP000007062 UP000075882 UP000007266 UP000261640 UP000075884
PRIDE
Pfam
PF00083 Sugar_tr
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JY31
A0A2H1V6U2
A0A2A4JCE7
A0A0N1PHI9
A0A0L7KA11
A0A2P8ZM01
+ More
A0A026WMM0 E2BT94 F4WBY5 V5GV83 A0A195BM51 A0A158NZ11 A0A026WP92 A0A3L8DXN6 A0A195FTD3 A0A3Q3E8J9 A0A3Q3EBD5 A0A151J628 A0A226EFB2 A0A151IEK9 A0A195FPC4 E0VNX8 A0A3Q3JG04 A0A3P8W6P3 A0A2G5BAM0 A0A3Q2C7A2 A0A1Y1MRS8 E2A9Z9 A0A084VRC5 A0A182MI44 Q7PR34 A0A182LFS7 D6X033 A0A3Q3KU25 A0A182N634
A0A026WMM0 E2BT94 F4WBY5 V5GV83 A0A195BM51 A0A158NZ11 A0A026WP92 A0A3L8DXN6 A0A195FTD3 A0A3Q3E8J9 A0A3Q3EBD5 A0A151J628 A0A226EFB2 A0A151IEK9 A0A195FPC4 E0VNX8 A0A3Q3JG04 A0A3P8W6P3 A0A2G5BAM0 A0A3Q2C7A2 A0A1Y1MRS8 E2A9Z9 A0A084VRC5 A0A182MI44 Q7PR34 A0A182LFS7 D6X033 A0A3Q3KU25 A0A182N634
PDB
4LDS
E-value=0.000275819,
Score=107
Ontologies
GO
Topology
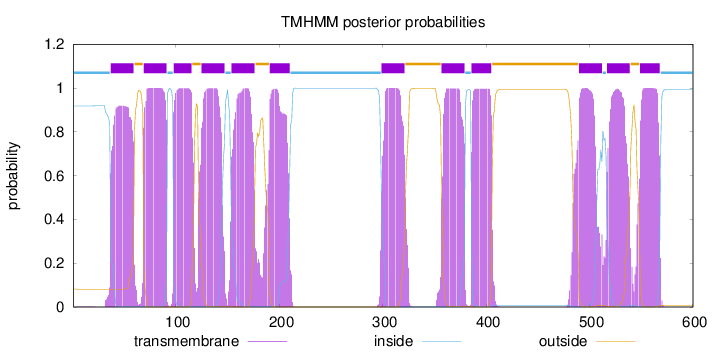
Length:
600
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
258.95261
Exp number, first 60 AAs:
20.29922
Total prob of N-in:
0.91789
POSSIBLE N-term signal
sequence
inside
1 - 36
TMhelix
37 - 59
outside
60 - 68
TMhelix
69 - 91
inside
92 - 97
TMhelix
98 - 115
outside
116 - 124
TMhelix
125 - 147
inside
148 - 153
TMhelix
154 - 176
outside
177 - 190
TMhelix
191 - 210
inside
211 - 298
TMhelix
299 - 321
outside
322 - 356
TMhelix
357 - 379
inside
380 - 385
TMhelix
386 - 405
outside
406 - 489
TMhelix
490 - 512
inside
513 - 516
TMhelix
517 - 539
outside
540 - 548
TMhelix
549 - 568
inside
569 - 600
Population Genetic Test Statistics
Pi
267.592633
Theta
178.71795
Tajima's D
1.583931
CLR
0.136277
CSRT
0.807559622018899
Interpretation
Uncertain
