Gene
KWMTBOMO11827
Pre Gene Modal
BGIBMGA013687
Annotation
chorion_specific_C/EBP_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 4.643
Sequence
CDS
ATGTACGATGCGGCGGCAGCGCCTCCGCCGCCTCCCCAACCAGACCTCAAAAAAGTCGTCGAGGATAAAAGATCGGCTTTCCCACCACCGGAACTCGACGAGCTCAATGGACAGGAGATCAGCTTGGATCTGCAACACCTCATCGAGGATCAGTTCCGGGGCGAGGAAACGATGGCCCTCTTTCAGGAAATCCTACCCGGAGGTCGATCGCCTCAGCCCCGGTTCACGAGGACGACGCTGGCGTACATGCCGCAACCGGTCCACTCCGGAGCGTCCTACGCGCCAGTACAAGCCAGCTCGGCACACGAGCAGGCGCCCCCAATTAAAGAAGAGCCGCCAGAACCGCAGGACTTCCGAAGAACGGTCACATGCTCGCAATACACAGGACAATACAATCCGCAGCCGCCAGTCGGCGTTAACAATCCTTACACAGGAAGTTTCACACCCCTACCGCCTCTGGGAGGACCTCTGCTTCCTCCTCTGCTGAAACACAAACCGGCACCTCCGAGACGATCCTCAGGCAAAGTAATAGACAAGGGAACTGACGAATACAGAAGGCGAAGGGAGCGCAACAACATAGCCGTGAGGAAATCCCGGGAAAAGGCTAAAGTACGTTCCCGCGAAGTCGAAGAAAAAGTGAAAACATTGCTGAGAGAGAAGGAGGCCTTGCTGAAGAGGCTCGAGGCGGTTTCGGGGGAGCTGAGCCTCCACAAGCAGATGTACGTGCACCTGATAAACTTGAACCACCCGGAGATCACGGAGCTGTGCCGGTCGATGCTGCAGCTGGGAGGCCCGCACTCCCAGGACCACACGCTTTGA
Protein
MYDAAAAPPPPPQPDLKKVVEDKRSAFPPPELDELNGQEISLDLQHLIEDQFRGEETMALFQEILPGGRSPQPRFTRTTLAYMPQPVHSGASYAPVQASSAHEQAPPIKEEPPEPQDFRRTVTCSQYTGQYNPQPPVGVNNPYTGSFTPLPPLGGPLLPPLLKHKPAPPRRSSGKVIDKGTDEYRRRRERNNIAVRKSREKAKVRSREVEEKVKTLLREKEALLKRLEAVSGELSLHKQMYVHLINLNHPEITELCRSMLQLGGPHSQDHTL
Summary
Uniprot
Q38Q34
A0A2A4K6V9
A0A3S2M5X0
A0A2H1WIZ2
A0A0N1PJV0
A0A194Q6Z9
+ More
A0A212F933 A0A0L7LLB3 A0A1B0DI02 A0A1B0GJ60 A0A1L8DIT6 Q7PSQ9 B0W7F3 A0A182KDS2 A0A336K8F1 Q17GX7 A0A1S4F2Y9 A0A182H8I9 A0A0K8TSC9 A0A0A1XN04 A0A1A9X2F9 A0A1B0G7V5 W8BPK2 A0A1A9XXV0 A0A1B0ADD0 A0A1A9VQ79 B4QBX4 A0A0K8U6A7 A0A1W4X310 A0A0L0BTR9 T1H5Y4 D6WSV6 E2B0S3 A0A1B6MMJ6 A0A1B6H8G7 A0A1B6GNM0 K7J0I8 A0A1Y1KJI8 A0A0J7NFA2 A0A026WYG6 A0A151IMJ2 A0A310SU83 A0A087ZZ61 A0A2S2NEY0 A0A2H8TYD4 A0A158NLI8 N6TT35 A0A195B1H0 A0A0L7RDM0 A0A195E6D7 E9I8R9 A0A2A3EIF4 J9JUE9 A0A154PQF5 A0A0A9YSI6 A0A3Q0IS76 E2BGL0 A0A146KXF4 F4WW11 A0A2J7R5X4
A0A212F933 A0A0L7LLB3 A0A1B0DI02 A0A1B0GJ60 A0A1L8DIT6 Q7PSQ9 B0W7F3 A0A182KDS2 A0A336K8F1 Q17GX7 A0A1S4F2Y9 A0A182H8I9 A0A0K8TSC9 A0A0A1XN04 A0A1A9X2F9 A0A1B0G7V5 W8BPK2 A0A1A9XXV0 A0A1B0ADD0 A0A1A9VQ79 B4QBX4 A0A0K8U6A7 A0A1W4X310 A0A0L0BTR9 T1H5Y4 D6WSV6 E2B0S3 A0A1B6MMJ6 A0A1B6H8G7 A0A1B6GNM0 K7J0I8 A0A1Y1KJI8 A0A0J7NFA2 A0A026WYG6 A0A151IMJ2 A0A310SU83 A0A087ZZ61 A0A2S2NEY0 A0A2H8TYD4 A0A158NLI8 N6TT35 A0A195B1H0 A0A0L7RDM0 A0A195E6D7 E9I8R9 A0A2A3EIF4 J9JUE9 A0A154PQF5 A0A0A9YSI6 A0A3Q0IS76 E2BGL0 A0A146KXF4 F4WW11 A0A2J7R5X4
Pubmed
EMBL
BABH01040620
DQ149981
DQ149982
AAZ84929.1
NWSH01000066
PCG80005.1
+ More
RSAL01000025 RVE52111.1 ODYU01008648 SOQ52414.1 KQ459838 KPJ19744.1 KQ459460 KPJ00775.1 AGBW02009650 OWR50252.1 JTDY01000647 KOB76348.1 AJVK01015435 AJWK01019388 GFDF01007814 JAV06270.1 AAAB01008816 EAA05185.5 DS231853 EDS37868.1 UFQS01000175 UFQT01000175 SSX00761.1 SSX21141.1 CH477254 EAT45947.1 JXUM01029872 KQ560866 KXJ80566.1 GDAI01000763 JAI16840.1 GBXI01002037 JAD12255.1 CCAG010021503 GAMC01007897 JAB98658.1 CM000362 EDX08529.1 GDHF01030122 JAI22192.1 JRES01001473 KNC22629.1 KQ971352 EFA06345.2 GL444666 EFN60745.1 GEBQ01002888 JAT37089.1 GECU01036775 GECU01025078 JAS70931.1 JAS82628.1 GECZ01005745 JAS64024.1 GEZM01081908 JAV61484.1 LBMM01005782 KMQ91210.1 KK107063 QOIP01000005 EZA61077.1 RLU22956.1 KQ977054 KYN06083.1 KQ760243 OAD61272.1 GGMR01003134 MBY15753.1 GFXV01007452 MBW19257.1 ADTU01019473 APGK01055478 KB741266 KB632194 ENN71506.1 ERL89938.1 KQ976662 KYM78338.1 KQ414613 KOC69087.1 KQ979579 KYN20651.1 GL761646 EFZ23032.1 KZ288231 PBC31565.1 ABLF02035570 KQ435037 KZC14146.1 GBHO01008475 GDHC01013126 JAG35129.1 JAQ05503.1 GL448189 EFN85203.1 GDHC01017485 JAQ01144.1 GL888404 EGI61600.1 NEVH01006987 PNF36238.1
RSAL01000025 RVE52111.1 ODYU01008648 SOQ52414.1 KQ459838 KPJ19744.1 KQ459460 KPJ00775.1 AGBW02009650 OWR50252.1 JTDY01000647 KOB76348.1 AJVK01015435 AJWK01019388 GFDF01007814 JAV06270.1 AAAB01008816 EAA05185.5 DS231853 EDS37868.1 UFQS01000175 UFQT01000175 SSX00761.1 SSX21141.1 CH477254 EAT45947.1 JXUM01029872 KQ560866 KXJ80566.1 GDAI01000763 JAI16840.1 GBXI01002037 JAD12255.1 CCAG010021503 GAMC01007897 JAB98658.1 CM000362 EDX08529.1 GDHF01030122 JAI22192.1 JRES01001473 KNC22629.1 KQ971352 EFA06345.2 GL444666 EFN60745.1 GEBQ01002888 JAT37089.1 GECU01036775 GECU01025078 JAS70931.1 JAS82628.1 GECZ01005745 JAS64024.1 GEZM01081908 JAV61484.1 LBMM01005782 KMQ91210.1 KK107063 QOIP01000005 EZA61077.1 RLU22956.1 KQ977054 KYN06083.1 KQ760243 OAD61272.1 GGMR01003134 MBY15753.1 GFXV01007452 MBW19257.1 ADTU01019473 APGK01055478 KB741266 KB632194 ENN71506.1 ERL89938.1 KQ976662 KYM78338.1 KQ414613 KOC69087.1 KQ979579 KYN20651.1 GL761646 EFZ23032.1 KZ288231 PBC31565.1 ABLF02035570 KQ435037 KZC14146.1 GBHO01008475 GDHC01013126 JAG35129.1 JAQ05503.1 GL448189 EFN85203.1 GDHC01017485 JAQ01144.1 GL888404 EGI61600.1 NEVH01006987 PNF36238.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000053268
UP000007151
+ More
UP000037510 UP000092462 UP000092461 UP000007062 UP000002320 UP000075881 UP000008820 UP000069940 UP000249989 UP000091820 UP000092444 UP000092443 UP000092445 UP000078200 UP000000304 UP000192223 UP000037069 UP000015102 UP000007266 UP000000311 UP000002358 UP000036403 UP000053097 UP000279307 UP000078542 UP000005203 UP000005205 UP000019118 UP000030742 UP000078540 UP000053825 UP000078492 UP000242457 UP000007819 UP000076502 UP000079169 UP000008237 UP000007755 UP000235965
UP000037510 UP000092462 UP000092461 UP000007062 UP000002320 UP000075881 UP000008820 UP000069940 UP000249989 UP000091820 UP000092444 UP000092443 UP000092445 UP000078200 UP000000304 UP000192223 UP000037069 UP000015102 UP000007266 UP000000311 UP000002358 UP000036403 UP000053097 UP000279307 UP000078542 UP000005203 UP000005205 UP000019118 UP000030742 UP000078540 UP000053825 UP000078492 UP000242457 UP000007819 UP000076502 UP000079169 UP000008237 UP000007755 UP000235965
PRIDE
Pfam
PF07716 bZIP_2
Interpro
IPR004827
bZIP
ProteinModelPortal
Q38Q34
A0A2A4K6V9
A0A3S2M5X0
A0A2H1WIZ2
A0A0N1PJV0
A0A194Q6Z9
+ More
A0A212F933 A0A0L7LLB3 A0A1B0DI02 A0A1B0GJ60 A0A1L8DIT6 Q7PSQ9 B0W7F3 A0A182KDS2 A0A336K8F1 Q17GX7 A0A1S4F2Y9 A0A182H8I9 A0A0K8TSC9 A0A0A1XN04 A0A1A9X2F9 A0A1B0G7V5 W8BPK2 A0A1A9XXV0 A0A1B0ADD0 A0A1A9VQ79 B4QBX4 A0A0K8U6A7 A0A1W4X310 A0A0L0BTR9 T1H5Y4 D6WSV6 E2B0S3 A0A1B6MMJ6 A0A1B6H8G7 A0A1B6GNM0 K7J0I8 A0A1Y1KJI8 A0A0J7NFA2 A0A026WYG6 A0A151IMJ2 A0A310SU83 A0A087ZZ61 A0A2S2NEY0 A0A2H8TYD4 A0A158NLI8 N6TT35 A0A195B1H0 A0A0L7RDM0 A0A195E6D7 E9I8R9 A0A2A3EIF4 J9JUE9 A0A154PQF5 A0A0A9YSI6 A0A3Q0IS76 E2BGL0 A0A146KXF4 F4WW11 A0A2J7R5X4
A0A212F933 A0A0L7LLB3 A0A1B0DI02 A0A1B0GJ60 A0A1L8DIT6 Q7PSQ9 B0W7F3 A0A182KDS2 A0A336K8F1 Q17GX7 A0A1S4F2Y9 A0A182H8I9 A0A0K8TSC9 A0A0A1XN04 A0A1A9X2F9 A0A1B0G7V5 W8BPK2 A0A1A9XXV0 A0A1B0ADD0 A0A1A9VQ79 B4QBX4 A0A0K8U6A7 A0A1W4X310 A0A0L0BTR9 T1H5Y4 D6WSV6 E2B0S3 A0A1B6MMJ6 A0A1B6H8G7 A0A1B6GNM0 K7J0I8 A0A1Y1KJI8 A0A0J7NFA2 A0A026WYG6 A0A151IMJ2 A0A310SU83 A0A087ZZ61 A0A2S2NEY0 A0A2H8TYD4 A0A158NLI8 N6TT35 A0A195B1H0 A0A0L7RDM0 A0A195E6D7 E9I8R9 A0A2A3EIF4 J9JUE9 A0A154PQF5 A0A0A9YSI6 A0A3Q0IS76 E2BGL0 A0A146KXF4 F4WW11 A0A2J7R5X4
PDB
1HJB
E-value=7.41102e-05,
Score=108
Ontologies
GO
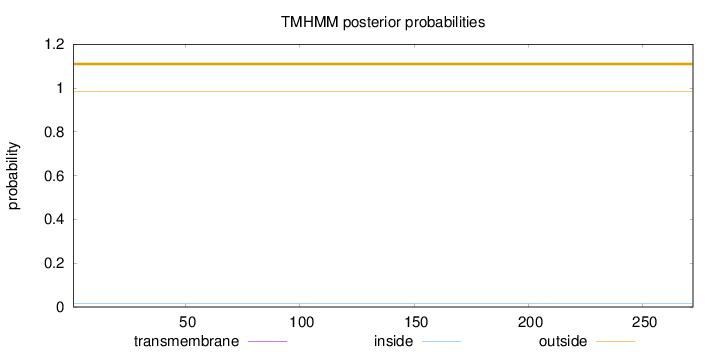
Topology
Length:
272
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00044
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01462
outside
1 - 272
Population Genetic Test Statistics
Pi
193.94257
Theta
172.266096
Tajima's D
0.369718
CLR
0.41399
CSRT
0.477426128693565
Interpretation
Uncertain
