Gene
KWMTBOMO11824
Pre Gene Modal
BGIBMGA013690
Annotation
PREDICTED:_ionotropic_GABA-aminobutyric_acid_receptor_RDL3_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.336
Sequence
CDS
ATGAGCGCCGTGCGTCACGCCGCGCTTCTCCTCGCGCTTGCCGCCACCTTCTTGCCGCAGGCCAATCGTGTCGCCGGCGGCGGTGGAGGTGGAATGTTCGGTGATGTCAATATATCAGCTATCTTGGATTCGTTCAGCATCAGCTATGATAAGCGAGTAAGACCAAACTATGGAGGTCCCCCCGTGGAAGTCGGCGTCACCATGTACGTCCTGTCCATCAGCTCCGTGTCTGAAGTCCTCATGGACTTCACTCTGGACTTCTACTTCAGACAGTTCTGGACAGATCCGCGACTGGCCTACAAGAAGCGACCAGGCGTGGAAACCCTGTCCGTGGGCTCGGAGTTCATCAAGAACATCTGGGTACCGGACACGTTCTTTGTCAACGAAAAGCAATCGTATTTCCACATAGCCACAACGAGCAACGAATTTATTAGGATTCACCACTCCGGCTCGATAACAAGGAGTATTAGATTAACGATCACTGCGTCATGTCCCATGAACCTGCAGTATTTCCCCATGGACCGGCAGCTTTGCCACATCGAGATTGAAAGCTTCGGCTACACGATGCGCGACATCCGCTACAAGTGGAACGAGGGACCCAACTCGGTGGGCGTCTCCAAGGAAGTGTCGCTGCCGCAGTTCAAGGTGCTCGGACACCGCCAGCGCGCCATGGAGATCTCGCTCACAACAGGGAATTACTCAAGGCTGGCGTGCGAAATACAGTTCGTACGCTCGATGGGGTACTACCTCATCCAGATCTACATACCTTCGGGTCTGATCGTGATCATATCGTGGGTCTCGTTCTGGCTGAACCGGAACGCGACCCCGGCCCGCGTGCAGCTAGGGGTCACCACCGTGCTGACGATGACAACGCTCATGTCATCGACCAACGCCGCTCTGCCGAAGATATCATACGTCAAGTCTATCGACGTGTACCTCGGGACCTGCTTCGTCATGGTTTTCACGAGCTTGCTTGAATACGCGACAGTTGGCTACATGTCGAAGAGGATACAGATGCGCAAGCAACGGTTCATTACCATACAAAAGATTATGGCCGAGAAGAAGATGCCCACGGAGTGTCCCCCGCCGTGGGACCCGCACACGCTCGCTAAAATGAGCTCCTTAAACCGCTACCCTCCTGGTAGCAGAGCTTCCGTGAGTAGAAGTTCGCAGGAAGTTCGCTACAAGGTCCGAGATCCGAAAGCGCACTCTAAGGGCGGCACGCTTGAGAACACGATCAGCAGGAGCAGGATGAACATCGAGGGGGACGCGGCGGGGCCCGTCATGCACCCCCTGCACCCGGGGAAGGACATTGGAAAATTCCTCGGCATGACGCCCTCGGACATCGACAAGTACTCGAGGATCGTGTTCCCGGTGTGCTTCGTCTGTTTCAACCTGGTCTACTGGATCGTGTACCTCCACGTGTCCGACGTGGTCGCCGAGGACCTGGAACTGCTCGAGGACAAGTGA
Protein
MSAVRHAALLLALAATFLPQANRVAGGGGGGMFGDVNISAILDSFSISYDKRVRPNYGGPPVEVGVTMYVLSISSVSEVLMDFTLDFYFRQFWTDPRLAYKKRPGVETLSVGSEFIKNIWVPDTFFVNEKQSYFHIATTSNEFIRIHHSGSITRSIRLTITASCPMNLQYFPMDRQLCHIEIESFGYTMRDIRYKWNEGPNSVGVSKEVSLPQFKVLGHRQRAMEISLTTGNYSRLACEIQFVRSMGYYLIQIYIPSGLIVIISWVSFWLNRNATPARVQLGVTTVLTMTTLMSSTNAALPKISYVKSIDVYLGTCFVMVFTSLLEYATVGYMSKRIQMRKQRFITIQKIMAEKKMPTECPPPWDPHTLAKMSSLNRYPPGSRASVSRSSQEVRYKVRDPKAHSKGGTLENTISRSRMNIEGDAAGPVMHPLHPGKDIGKFLGMTPSDIDKYSRIVFPVCFVCFNLVYWIVYLHVSDVVAEDLELLEDK
Summary
Similarity
Belongs to the ligand-gated ion channel (TC 1.A.9) family.
Uniprot
E0X9I2
E0X9I4
E0X9I1
H9JVX7
E0X9I3
A0A3G1J4Z2
+ More
O18471 A0A2S0DGE1 E0X9I5 A0A1Q2T844 A0A0U5ACA3 U3TMA1 E0X9H7 A5A2M7 C0L7I4 A0A2H1WJ59 A9LYI3 C0L7I5 A0A3S2NX06 A0A1Q2T857 A0A2A4JF95 E0X9H8 E0X9H9 C0L956 A1Z0K6 A3FBG6 C0L955 A0A1Q2T851 G8IIS8 O18468 V9ZCF2 V9Z984 Q9XZW0 A5JJ42 A0A1Q2T839 A0A2A4JFC5 A0A1X9PRB2 H3JU08 A0A2J7QNV6 A8DMU0 A0A1W4WF43 M9Z6B1 A0A1W4WE60 A0A168SJQ0 A0A1I9VT68 E2IKR3 A8DMU1 A0A087Z8M8 G8IIR9 A0A168SJP2 A0A1Q2T841 A0A1I8JSK2 Q0GQR7 A0A182QGF5 A0A1W4W3J7 A0A1W4WE55 A0A182XM21 A0A182URJ2 A0A1Y9GKS7 A7UU05 A0A1Y9HEH3 A0A182J2V0 A0A158NFE0 A0A2H4WAU8 A0A0B5EEN9 A0A1I8QEE6 A0A168SJJ0 X5C002 X5BIT8 A0A1Y9H213 A0A1Y9IVD6 A0A1I8QEJ2 A0A0Q9WTB8 A0A2A4JL08 A0A2J7QNV0 A0A0K8STF5 A0A0K8TDA0 C0L7I6 A0A1Q2T840 A0A1J1J944 B4LHG7 A0A0Q9WHF3 F5C3U5 A0A1I8QED8 A0A0Q9WGK3 A0A2W1BQH7 A0A1I8QEF2 C0L7I7 A0A2J7QNV2 A8DMT9 A0A0M4EK86 A0A1B6DGU4 K7ZRK2 O17145 A0A0Q9XB93 A0A1W4WFF3 A0A0Q9WH84 B4KWM3 A0A1Q2T843 A0A1I8M4J7
O18471 A0A2S0DGE1 E0X9I5 A0A1Q2T844 A0A0U5ACA3 U3TMA1 E0X9H7 A5A2M7 C0L7I4 A0A2H1WJ59 A9LYI3 C0L7I5 A0A3S2NX06 A0A1Q2T857 A0A2A4JF95 E0X9H8 E0X9H9 C0L956 A1Z0K6 A3FBG6 C0L955 A0A1Q2T851 G8IIS8 O18468 V9ZCF2 V9Z984 Q9XZW0 A5JJ42 A0A1Q2T839 A0A2A4JFC5 A0A1X9PRB2 H3JU08 A0A2J7QNV6 A8DMU0 A0A1W4WF43 M9Z6B1 A0A1W4WE60 A0A168SJQ0 A0A1I9VT68 E2IKR3 A8DMU1 A0A087Z8M8 G8IIR9 A0A168SJP2 A0A1Q2T841 A0A1I8JSK2 Q0GQR7 A0A182QGF5 A0A1W4W3J7 A0A1W4WE55 A0A182XM21 A0A182URJ2 A0A1Y9GKS7 A7UU05 A0A1Y9HEH3 A0A182J2V0 A0A158NFE0 A0A2H4WAU8 A0A0B5EEN9 A0A1I8QEE6 A0A168SJJ0 X5C002 X5BIT8 A0A1Y9H213 A0A1Y9IVD6 A0A1I8QEJ2 A0A0Q9WTB8 A0A2A4JL08 A0A2J7QNV0 A0A0K8STF5 A0A0K8TDA0 C0L7I6 A0A1Q2T840 A0A1J1J944 B4LHG7 A0A0Q9WHF3 F5C3U5 A0A1I8QED8 A0A0Q9WGK3 A0A2W1BQH7 A0A1I8QEF2 C0L7I7 A0A2J7QNV2 A8DMT9 A0A0M4EK86 A0A1B6DGU4 K7ZRK2 O17145 A0A0Q9XB93 A0A1W4WFF3 A0A0Q9WH84 B4KWM3 A0A1Q2T843 A0A1I8M4J7
Pubmed
EMBL
GQ890666
ADM88011.1
GQ890668
ADM88013.1
GQ890665
ADM88010.1
+ More
BABH01040612 GQ890667 ADM88012.1 KX856966 ASY91958.1 AF006192 AAB62572.1 KX856969 ASY91961.1 GQ890669 ADM88014.1 LC171477 BAW87784.1 AB278155 BAT57340.1 AB847423 BAN92442.1 GQ890661 ADM88006.1 EF521821 ABP96889.1 FJ665607 ACN52597.1 ODYU01009008 SOQ53088.1 EU273945 ABX47168.1 FJ665608 ACN52598.1 RSAL01000121 RVE46688.1 LC171476 BAW87783.1 NWSH01001799 PCG70103.1 GQ890662 ADM88007.1 GQ890663 ADM88008.1 FJ668709 ACN53549.1 EF156251 ABL86443.1 EF368369 ABN13426.1 ACN53550.1 LC171475 BAW87782.1 JN794059 AER59668.1 AF006189 AAB62563.1 KF881790 AHE41087.1 KF881791 AHE41088.1 AJ224513 CAB41615.1 EF535530 ABQ45398.1 LC171472 BAW87779.1 PCG70102.1 KY072798 ARO86294.1 AB617631 BAL63029.1 NEVH01012562 PNF30266.1 EF545117 KQ972204 ABU63595.1 KYB24596.1 KC841916 AGK30293.1 KU201323 ANC68178.1 KX431144 APA16887.1 HM566200 KX856970 ADK74004.1 ASY91962.1 EF545118 ABU63596.1 KYB24597.1 JN792582 AER59667.1 KU201322 ANC68177.1 LC171474 BAW87781.1 DQ667182 ABG75734.1 AXCN02000404 APCN01000072 APCN01000073 AAAB01008960 EDO63781.1 ADTU01014314 ADTU01014315 ADTU01014316 ADTU01014317 ADTU01014318 ADTU01014319 ADTU01014320 ADTU01014321 MG190358 AUC64171.1 KJ485710 AJE68941.1 KU201321 ANC68176.1 KJ174465 AHW29555.1 KJ174466 AHW29556.1 CH940647 KRF83883.1 NWSH01001075 PCG72737.1 PNF30265.1 GBRD01009258 JAG56563.1 GBRD01002284 JAG63537.1 FJ665609 ACN52599.1 LC171471 BAW87778.1 CVRI01000075 CRL08578.1 EDW68497.2 KRF83882.1 JF460792 AEB60992.1 KRF83884.1 KZ149977 PZC75884.1 FJ665610 ACN52600.1 PNF30267.1 EF545116 ABU63594.1 EFA12941.2 CP012525 ALC43610.1 GEDC01012391 JAS24907.1 AB697765 BAM66322.1 AF024647 AAB81966.1 CH933809 KRG05565.1 KRF83878.1 KRF83879.1 KRF83880.1 KRF83881.1 EDW17470.2 LC171473 BAW87780.1
BABH01040612 GQ890667 ADM88012.1 KX856966 ASY91958.1 AF006192 AAB62572.1 KX856969 ASY91961.1 GQ890669 ADM88014.1 LC171477 BAW87784.1 AB278155 BAT57340.1 AB847423 BAN92442.1 GQ890661 ADM88006.1 EF521821 ABP96889.1 FJ665607 ACN52597.1 ODYU01009008 SOQ53088.1 EU273945 ABX47168.1 FJ665608 ACN52598.1 RSAL01000121 RVE46688.1 LC171476 BAW87783.1 NWSH01001799 PCG70103.1 GQ890662 ADM88007.1 GQ890663 ADM88008.1 FJ668709 ACN53549.1 EF156251 ABL86443.1 EF368369 ABN13426.1 ACN53550.1 LC171475 BAW87782.1 JN794059 AER59668.1 AF006189 AAB62563.1 KF881790 AHE41087.1 KF881791 AHE41088.1 AJ224513 CAB41615.1 EF535530 ABQ45398.1 LC171472 BAW87779.1 PCG70102.1 KY072798 ARO86294.1 AB617631 BAL63029.1 NEVH01012562 PNF30266.1 EF545117 KQ972204 ABU63595.1 KYB24596.1 KC841916 AGK30293.1 KU201323 ANC68178.1 KX431144 APA16887.1 HM566200 KX856970 ADK74004.1 ASY91962.1 EF545118 ABU63596.1 KYB24597.1 JN792582 AER59667.1 KU201322 ANC68177.1 LC171474 BAW87781.1 DQ667182 ABG75734.1 AXCN02000404 APCN01000072 APCN01000073 AAAB01008960 EDO63781.1 ADTU01014314 ADTU01014315 ADTU01014316 ADTU01014317 ADTU01014318 ADTU01014319 ADTU01014320 ADTU01014321 MG190358 AUC64171.1 KJ485710 AJE68941.1 KU201321 ANC68176.1 KJ174465 AHW29555.1 KJ174466 AHW29556.1 CH940647 KRF83883.1 NWSH01001075 PCG72737.1 PNF30265.1 GBRD01009258 JAG56563.1 GBRD01002284 JAG63537.1 FJ665609 ACN52599.1 LC171471 BAW87778.1 CVRI01000075 CRL08578.1 EDW68497.2 KRF83882.1 JF460792 AEB60992.1 KRF83884.1 KZ149977 PZC75884.1 FJ665610 ACN52600.1 PNF30267.1 EF545116 ABU63594.1 EFA12941.2 CP012525 ALC43610.1 GEDC01012391 JAS24907.1 AB697765 BAM66322.1 AF024647 AAB81966.1 CH933809 KRG05565.1 KRF83878.1 KRF83879.1 KRF83880.1 KRF83881.1 EDW17470.2 LC171473 BAW87780.1
Proteomes
Interpro
Gene 3D
ProteinModelPortal
E0X9I2
E0X9I4
E0X9I1
H9JVX7
E0X9I3
A0A3G1J4Z2
+ More
O18471 A0A2S0DGE1 E0X9I5 A0A1Q2T844 A0A0U5ACA3 U3TMA1 E0X9H7 A5A2M7 C0L7I4 A0A2H1WJ59 A9LYI3 C0L7I5 A0A3S2NX06 A0A1Q2T857 A0A2A4JF95 E0X9H8 E0X9H9 C0L956 A1Z0K6 A3FBG6 C0L955 A0A1Q2T851 G8IIS8 O18468 V9ZCF2 V9Z984 Q9XZW0 A5JJ42 A0A1Q2T839 A0A2A4JFC5 A0A1X9PRB2 H3JU08 A0A2J7QNV6 A8DMU0 A0A1W4WF43 M9Z6B1 A0A1W4WE60 A0A168SJQ0 A0A1I9VT68 E2IKR3 A8DMU1 A0A087Z8M8 G8IIR9 A0A168SJP2 A0A1Q2T841 A0A1I8JSK2 Q0GQR7 A0A182QGF5 A0A1W4W3J7 A0A1W4WE55 A0A182XM21 A0A182URJ2 A0A1Y9GKS7 A7UU05 A0A1Y9HEH3 A0A182J2V0 A0A158NFE0 A0A2H4WAU8 A0A0B5EEN9 A0A1I8QEE6 A0A168SJJ0 X5C002 X5BIT8 A0A1Y9H213 A0A1Y9IVD6 A0A1I8QEJ2 A0A0Q9WTB8 A0A2A4JL08 A0A2J7QNV0 A0A0K8STF5 A0A0K8TDA0 C0L7I6 A0A1Q2T840 A0A1J1J944 B4LHG7 A0A0Q9WHF3 F5C3U5 A0A1I8QED8 A0A0Q9WGK3 A0A2W1BQH7 A0A1I8QEF2 C0L7I7 A0A2J7QNV2 A8DMT9 A0A0M4EK86 A0A1B6DGU4 K7ZRK2 O17145 A0A0Q9XB93 A0A1W4WFF3 A0A0Q9WH84 B4KWM3 A0A1Q2T843 A0A1I8M4J7
O18471 A0A2S0DGE1 E0X9I5 A0A1Q2T844 A0A0U5ACA3 U3TMA1 E0X9H7 A5A2M7 C0L7I4 A0A2H1WJ59 A9LYI3 C0L7I5 A0A3S2NX06 A0A1Q2T857 A0A2A4JF95 E0X9H8 E0X9H9 C0L956 A1Z0K6 A3FBG6 C0L955 A0A1Q2T851 G8IIS8 O18468 V9ZCF2 V9Z984 Q9XZW0 A5JJ42 A0A1Q2T839 A0A2A4JFC5 A0A1X9PRB2 H3JU08 A0A2J7QNV6 A8DMU0 A0A1W4WF43 M9Z6B1 A0A1W4WE60 A0A168SJQ0 A0A1I9VT68 E2IKR3 A8DMU1 A0A087Z8M8 G8IIR9 A0A168SJP2 A0A1Q2T841 A0A1I8JSK2 Q0GQR7 A0A182QGF5 A0A1W4W3J7 A0A1W4WE55 A0A182XM21 A0A182URJ2 A0A1Y9GKS7 A7UU05 A0A1Y9HEH3 A0A182J2V0 A0A158NFE0 A0A2H4WAU8 A0A0B5EEN9 A0A1I8QEE6 A0A168SJJ0 X5C002 X5BIT8 A0A1Y9H213 A0A1Y9IVD6 A0A1I8QEJ2 A0A0Q9WTB8 A0A2A4JL08 A0A2J7QNV0 A0A0K8STF5 A0A0K8TDA0 C0L7I6 A0A1Q2T840 A0A1J1J944 B4LHG7 A0A0Q9WHF3 F5C3U5 A0A1I8QED8 A0A0Q9WGK3 A0A2W1BQH7 A0A1I8QEF2 C0L7I7 A0A2J7QNV2 A8DMT9 A0A0M4EK86 A0A1B6DGU4 K7ZRK2 O17145 A0A0Q9XB93 A0A1W4WFF3 A0A0Q9WH84 B4KWM3 A0A1Q2T843 A0A1I8M4J7
PDB
6I53
E-value=3.41857e-78,
Score=743
Ontologies
GO
PANTHER
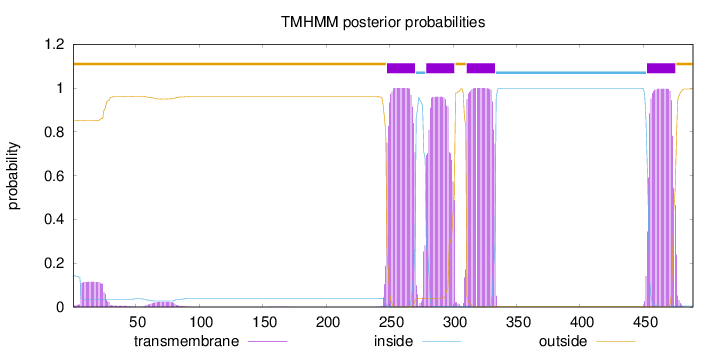
Topology
SignalP
Position: 1 - 26,
Likelihood: 0.968734

Length:
489
Number of predicted TMHs:
4
Exp number of AAs in TMHs:
89.21974
Exp number, first 60 AAs:
2.40561
Total prob of N-in:
0.14808
outside
1 - 247
TMhelix
248 - 270
inside
271 - 278
TMhelix
279 - 301
outside
302 - 310
TMhelix
311 - 333
inside
334 - 452
TMhelix
453 - 475
outside
476 - 489
Population Genetic Test Statistics
Pi
170.237433
Theta
156.691549
Tajima's D
0.37008
CLR
0.422128
CSRT
0.47607619619019
Interpretation
Uncertain
