Gene
KWMTBOMO11821
Annotation
PREDICTED:_chondroitin_sulfate_synthase_2_[Amyelois_transitella]
Full name
Hexosyltransferase
Location in the cell
Extracellular Reliability : 1.11
Sequence
CDS
ATGTTCTCGCGATGCCTGGTGTCTCAAGTAAAACATAACTCGTACTTTTTAATAGGCCTATTAATCGGTTTGTGGGTGTCTCTCGCTGTTATACCGTTGGATGAAGAGCCGGCGCCTTGTGTAGCGACTGTAGGCAGTGCCTCTCCAGTTCAGGATGAATTTGTGCCGCAGCGGGAGGAAAAACCAGCTGGTTCAGGACAGACGGCTGGGAGAAGCGTGCAACGACCTCGTTATTACAGTACTGAGCTGGGCATGAGAGGTTCCCTTCTCACTGGAATCCTGAGCTCGGAAGAAGCCCTCAAAGTCCAAGTGCCAGCTCTGAACCGAACAGCAGCAAGACTGCAGCCAGCCTTAAAATTCTTCATTACCGCCAGTGCAATGAGCACTGTACCTGGGCTAGCTAATGTTGTTGGCTTCACAGATACTAGGGAAATGCTAAAACCATTTCATGCCTTGAAGTACTTAGCAGATAATTATTTAGAGGAGTACGATTTCTTCTTCATTGTATCCGACACAGCCTTTGTGAATGCAAGACGGTTGACCCAGCTGGTCTCTCAGTTGTCTGTCAGCCAAGATGTGTACATGGGGAGGGTAGCTGATGATGAAAGTCATTATTGTTCTTTAGACAATGGAATAATCCTTTCTAACTCTGTGCTCCGCGCTATCCACAGTGAACTCGACTGGTGTGTACGCAATTCATATTCTCCTCACCACTACGAGAATATAGGTCGTTGTGTGTTACACTCTGCTCATTTGTCTTGCAACCCTGACGTACAGGGTGAACACTACTCATCAGTAGTTATCGAAAATGATCAACTACCATTAACTGAGTCTTTGGCTGATGCTGTAACAGTGTATCCCATCATGGACACTCACAGGTTCTACGAGATACACGCTTACATATCACGGGTGTTTTTGGAACGCGACCACGAGGAGGTGCTGAAACTACGCAGCTACATGTGGCACAACTCCGCGCGCCACCCCCCCGGGTACAGGAACAGCACGTGGCCCGCCGGCCTCAGGGACGATCCCGGACTGGCCAGACCTTTACCGGGGAACAGGTTCGACCACCTCCGCTGGACGGCAATTAACGGGACCCACGCGTTCTTTCCTGACGACCACCACAATGTATTACCGATCACCGGAGCTTACAGAGAAGCCTTGGATCTGGTGCTCACCCAAGTGAGGGCGTGGTCGCTGCACCGCTGGCCCGAGGCGCTGGACGTGCAGCTGGTGGAGGGGGCCTGGAAGTGGGAGCCGGCCGTCGCGCTGCGGTACCGCCTCCTGCTCCGCCTCACCGCCAAGGAGGACAAGGACACGCACATGAGCAGGCTGGTGGAGGCGGTGCGTCCGCTGGGCGCGGCGCGGCTGGTGCCGGTGCGCTACGTGACGGAGAGCGCGCGGCTCAGCGCCGTGCTGGCGGCGCCGCACACCGCGCGCCACGCCGCCGACCTCGCCGCCTTCCTGCAGCGGTACGAGGCGGTCTGCCTTAACCAGGACAAGAACACCGCGCTCCTGCTGGTGATAGTGAAGCAGCCTTCCCAGCCTTCCGTCGGCAACACCACTGAGGTGGACCTCATGAAGGAGAACAAGGAGACGGTGGCCGCGCTGAAGTCCCGGCACAAGGACGCCCAGATCGAGGTGGTCGAGACGAGCCTGGAGTCTCCCAGCCACGAGCACGCCAGCGAGGTGGAGCTGCTGCAGGAGGCGCAGCGGGCCGCGCTCGCCCTCGCCCTGCAGCGGGTACCCCGGGACAGCCTGGTGCTCCTCGCCGTGCCGCACATCGACTTCAACCAAGACTTCTTGAATCGGGTCCGCATGAACACGATATCCGGCGAGCAGTGGTACCTGCCCAGCGCGTTCTCCCGCTACGCGCAGTACGAGCACCCCGCCTTCGTGACGCCCACCGGCGCCAAGCCGCAGTCCAACACCGGCCGCTTCAACGTGCACCTCAGCCTCGTCTCCTCCTTCTACAGGAGCGACTACGACTCGGCGGTGGCGGAGTGGCCCGGGCCGGGGGGGGCGGACCCCGCGGCGGTGCTGGTCCGCAGCGGGCTGCGCTGCGTGCGCGCACCCGAGCCGGGGCTGGTGTACGCGTCGCGGGGGGCGCCGTGCGGGGGCGCGGGGGACGCGGCGTGCGTGTCGCGGCTGGTCACGCAGCTGCACACGCTGGACCTCGGCGCCAAGCACTCGCTCGCACAGCTGCTGCTCGAAACTCAAGCCGAGCTGGACCAGTAG
Protein
MFSRCLVSQVKHNSYFLIGLLIGLWVSLAVIPLDEEPAPCVATVGSASPVQDEFVPQREEKPAGSGQTAGRSVQRPRYYSTELGMRGSLLTGILSSEEALKVQVPALNRTAARLQPALKFFITASAMSTVPGLANVVGFTDTREMLKPFHALKYLADNYLEEYDFFFIVSDTAFVNARRLTQLVSQLSVSQDVYMGRVADDESHYCSLDNGIILSNSVLRAIHSELDWCVRNSYSPHHYENIGRCVLHSAHLSCNPDVQGEHYSSVVIENDQLPLTESLADAVTVYPIMDTHRFYEIHAYISRVFLERDHEEVLKLRSYMWHNSARHPPGYRNSTWPAGLRDDPGLARPLPGNRFDHLRWTAINGTHAFFPDDHHNVLPITGAYREALDLVLTQVRAWSLHRWPEALDVQLVEGAWKWEPAVALRYRLLLRLTAKEDKDTHMSRLVEAVRPLGAARLVPVRYVTESARLSAVLAAPHTARHAADLAAFLQRYEAVCLNQDKNTALLLVIVKQPSQPSVGNTTEVDLMKENKETVAALKSRHKDAQIEVVETSLESPSHEHASEVELLQEAQRAALALALQRVPRDSLVLLAVPHIDFNQDFLNRVRMNTISGEQWYLPSAFSRYAQYEHPAFVTPTGAKPQSNTGRFNVHLSLVSSFYRSDYDSAVAEWPGPGGADPAAVLVRSGLRCVRAPEPGLVYASRGAPCGGAGDAACVSRLVTQLHTLDLGAKHSLAQLLLETQAELDQ
Summary
Similarity
Belongs to the chondroitin N-acetylgalactosaminyltransferase family.
Feature
chain Hexosyltransferase
Uniprot
A0A2A4J776
A0A212EXY8
A0A2P8YIB7
K7IVJ9
A0A232EF39
A0A1S3CZ03
+ More
E2C7H7 A0A1B6C963 E2AAJ9 A0A3L8DY01 A0A026WLV0 A0A067QR13 A0A2J7Q9V5 A0A2J7Q9T6 A0A1S4KFX2 E9JDC0 J9K2F5 B0XG38 Q16SL4 A0A1S4FQP4 A0A158NZ16 A0A1Y1N8D2 A0A2R7VUX5 A0A2H8TN53 A0A139WGD2 A0A2S2QQV0 A0A2S2P464 A0A2A3EN76 A0A182FMM8 A0A182SW67 F4WBZ6 U5ER64 A0A0V0G6G6 A0A088A1X0 A0A151WQA1 A0A1L8E333 A0A1L8E3H9 A0A2J7Q9T2 A0A182H4Z9 A0A224XL69 A0A2M4BDU7 A0A195BMF5 A0A336K0T1 A0A2M4BDZ1 A0A2M4BDU6 A0A195FSY4 A0A2M3Z6S2 T1HXA0 A0A2M4ADF2 A0A0K8WM52 A0A151J2P1 A0A0C9QEI3 A0A182IYS0 A0A1J1IVP5 A0A0M3QZ51 A0A1W4VKV0 A0A146L6G1 A0A182W611 A0A1W4V7R0 A0A0A9YFR8 Q7PMP8 B3MS44 A0A0P4W3X9 A0A084VGQ1 A0A0L7QUQ3 Q9VYH2 A0A1W4VK35 Q29IB5 A0A1B6FWX5 A0A1B6L696 A0A182MHM8 A0A2J7Q9V0 B4M726 A0A1B0GL08 A0A087UVX6 A0A1B6J300 A0A1J1IXP3 N6TR11 A0A2P6K3S7 U4UIC4 V4AXE2 A0A1S3IB52 A0A210PSJ7 K1QTT0 S4RPI0 R0KYT2 R7TAR8 A0A087QXS9 A0A2I0U248 A0A2K6GEP2 A0A131XVR8 T1J8E4 B7Q2Y7
E2C7H7 A0A1B6C963 E2AAJ9 A0A3L8DY01 A0A026WLV0 A0A067QR13 A0A2J7Q9V5 A0A2J7Q9T6 A0A1S4KFX2 E9JDC0 J9K2F5 B0XG38 Q16SL4 A0A1S4FQP4 A0A158NZ16 A0A1Y1N8D2 A0A2R7VUX5 A0A2H8TN53 A0A139WGD2 A0A2S2QQV0 A0A2S2P464 A0A2A3EN76 A0A182FMM8 A0A182SW67 F4WBZ6 U5ER64 A0A0V0G6G6 A0A088A1X0 A0A151WQA1 A0A1L8E333 A0A1L8E3H9 A0A2J7Q9T2 A0A182H4Z9 A0A224XL69 A0A2M4BDU7 A0A195BMF5 A0A336K0T1 A0A2M4BDZ1 A0A2M4BDU6 A0A195FSY4 A0A2M3Z6S2 T1HXA0 A0A2M4ADF2 A0A0K8WM52 A0A151J2P1 A0A0C9QEI3 A0A182IYS0 A0A1J1IVP5 A0A0M3QZ51 A0A1W4VKV0 A0A146L6G1 A0A182W611 A0A1W4V7R0 A0A0A9YFR8 Q7PMP8 B3MS44 A0A0P4W3X9 A0A084VGQ1 A0A0L7QUQ3 Q9VYH2 A0A1W4VK35 Q29IB5 A0A1B6FWX5 A0A1B6L696 A0A182MHM8 A0A2J7Q9V0 B4M726 A0A1B0GL08 A0A087UVX6 A0A1B6J300 A0A1J1IXP3 N6TR11 A0A2P6K3S7 U4UIC4 V4AXE2 A0A1S3IB52 A0A210PSJ7 K1QTT0 S4RPI0 R0KYT2 R7TAR8 A0A087QXS9 A0A2I0U248 A0A2K6GEP2 A0A131XVR8 T1J8E4 B7Q2Y7
EC Number
2.4.1.-
Pubmed
22118469
29403074
20075255
28648823
20798317
30249741
+ More
24508170 24845553 21282665 17510324 21347285 28004739 18362917 19820115 21719571 26483478 26823975 25401762 12364791 14747013 17210077 17994087 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23537049 23254933 28812685 22992520
24508170 24845553 21282665 17510324 21347285 28004739 18362917 19820115 21719571 26483478 26823975 25401762 12364791 14747013 17210077 17994087 24438588 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 23537049 23254933 28812685 22992520
EMBL
NWSH01002628
PCG67811.1
AGBW02011687
OWR46304.1
PYGN01000578
PSN43934.1
+ More
NNAY01005141 OXU16953.1 GL453369 EFN76101.1 GEDC01027429 JAS09869.1 GL438128 EFN69546.1 QOIP01000003 RLU24985.1 KK107161 EZA56601.1 KK853123 KDR11049.1 NEVH01016344 PNF25349.1 PNF25348.1 GL771866 EFZ09144.1 ABLF02029482 DS232985 EDS27213.1 CH477672 EAT37450.1 ADTU01004505 GEZM01010916 JAV93728.1 KK854101 PTY11294.1 GFXV01002903 MBW14708.1 KQ971348 KYB26915.1 GGMS01010881 MBY80084.1 GGMR01011389 MBY24008.1 KZ288215 PBC32656.1 GL888067 EGI68285.1 GANO01002946 JAB56925.1 GECL01002836 JAP03288.1 KQ982837 KYQ50069.1 GFDF01000946 JAV13138.1 GFDF01000890 JAV13194.1 PNF25351.1 JXUM01110504 JXUM01110505 JXUM01110506 KQ565419 KXJ71000.1 GFTR01007657 JAW08769.1 GGFJ01002074 MBW51215.1 KQ976444 KYM86211.1 UFQS01000015 UFQT01000015 SSW97392.1 SSX17778.1 GGFJ01002072 MBW51213.1 GGFJ01002073 MBW51214.1 KQ981276 KYN43558.1 GGFM01003471 MBW24222.1 ACPB03006224 GGFK01005327 MBW38648.1 GDHF01000354 JAI51960.1 KQ980353 KYN16354.1 GBYB01001694 GBYB01007979 JAG71461.1 JAG77746.1 CVRI01000063 CRL04339.1 CP012528 ALC48759.1 GDHC01014656 JAQ03973.1 GBHO01011692 GBHO01011691 JAG31912.1 JAG31913.1 AAAB01008973 EAA13433.5 CH902622 EDV34599.2 GDRN01093994 JAI59779.1 ATLV01013047 KE524836 KFB37145.1 KQ414735 KOC62206.1 AE014298 AAF48225.2 CH379063 EAL32738.3 GECZ01015077 JAS54692.1 GEBQ01020953 JAT19024.1 AXCM01001949 PNF25352.1 CH940653 EDW62593.2 AJWK01034037 AJWK01034038 AJWK01034039 KK121911 KFM81515.1 GECU01014164 JAS93542.1 CRL04340.1 APGK01055312 KB741261 ENN71665.1 MWRG01032027 PRD20985.1 KB632310 ERL92113.1 KB200314 ESP02248.1 NEDP02005525 OWF39426.1 JH816951 EKC40242.1 KB744814 EOA94349.1 AMQN01003053 KB310836 ELT90607.1 KL225993 KFM06033.1 KZ506339 PKU40099.1 GEFM01005491 JAP70305.1 JH431954 ABJB010277392 DS847090 EEC13209.1
NNAY01005141 OXU16953.1 GL453369 EFN76101.1 GEDC01027429 JAS09869.1 GL438128 EFN69546.1 QOIP01000003 RLU24985.1 KK107161 EZA56601.1 KK853123 KDR11049.1 NEVH01016344 PNF25349.1 PNF25348.1 GL771866 EFZ09144.1 ABLF02029482 DS232985 EDS27213.1 CH477672 EAT37450.1 ADTU01004505 GEZM01010916 JAV93728.1 KK854101 PTY11294.1 GFXV01002903 MBW14708.1 KQ971348 KYB26915.1 GGMS01010881 MBY80084.1 GGMR01011389 MBY24008.1 KZ288215 PBC32656.1 GL888067 EGI68285.1 GANO01002946 JAB56925.1 GECL01002836 JAP03288.1 KQ982837 KYQ50069.1 GFDF01000946 JAV13138.1 GFDF01000890 JAV13194.1 PNF25351.1 JXUM01110504 JXUM01110505 JXUM01110506 KQ565419 KXJ71000.1 GFTR01007657 JAW08769.1 GGFJ01002074 MBW51215.1 KQ976444 KYM86211.1 UFQS01000015 UFQT01000015 SSW97392.1 SSX17778.1 GGFJ01002072 MBW51213.1 GGFJ01002073 MBW51214.1 KQ981276 KYN43558.1 GGFM01003471 MBW24222.1 ACPB03006224 GGFK01005327 MBW38648.1 GDHF01000354 JAI51960.1 KQ980353 KYN16354.1 GBYB01001694 GBYB01007979 JAG71461.1 JAG77746.1 CVRI01000063 CRL04339.1 CP012528 ALC48759.1 GDHC01014656 JAQ03973.1 GBHO01011692 GBHO01011691 JAG31912.1 JAG31913.1 AAAB01008973 EAA13433.5 CH902622 EDV34599.2 GDRN01093994 JAI59779.1 ATLV01013047 KE524836 KFB37145.1 KQ414735 KOC62206.1 AE014298 AAF48225.2 CH379063 EAL32738.3 GECZ01015077 JAS54692.1 GEBQ01020953 JAT19024.1 AXCM01001949 PNF25352.1 CH940653 EDW62593.2 AJWK01034037 AJWK01034038 AJWK01034039 KK121911 KFM81515.1 GECU01014164 JAS93542.1 CRL04340.1 APGK01055312 KB741261 ENN71665.1 MWRG01032027 PRD20985.1 KB632310 ERL92113.1 KB200314 ESP02248.1 NEDP02005525 OWF39426.1 JH816951 EKC40242.1 KB744814 EOA94349.1 AMQN01003053 KB310836 ELT90607.1 KL225993 KFM06033.1 KZ506339 PKU40099.1 GEFM01005491 JAP70305.1 JH431954 ABJB010277392 DS847090 EEC13209.1
Proteomes
UP000218220
UP000007151
UP000245037
UP000002358
UP000215335
UP000079169
+ More
UP000008237 UP000000311 UP000279307 UP000053097 UP000027135 UP000235965 UP000007819 UP000002320 UP000008820 UP000005205 UP000007266 UP000242457 UP000069272 UP000075901 UP000007755 UP000005203 UP000075809 UP000069940 UP000249989 UP000078540 UP000078541 UP000015103 UP000078492 UP000075880 UP000183832 UP000092553 UP000192221 UP000075920 UP000007062 UP000007801 UP000030765 UP000053825 UP000000803 UP000001819 UP000075883 UP000008792 UP000092461 UP000054359 UP000019118 UP000030742 UP000030746 UP000085678 UP000242188 UP000005408 UP000245300 UP000014760 UP000053286 UP000233160 UP000001555
UP000008237 UP000000311 UP000279307 UP000053097 UP000027135 UP000235965 UP000007819 UP000002320 UP000008820 UP000005205 UP000007266 UP000242457 UP000069272 UP000075901 UP000007755 UP000005203 UP000075809 UP000069940 UP000249989 UP000078540 UP000078541 UP000015103 UP000078492 UP000075880 UP000183832 UP000092553 UP000192221 UP000075920 UP000007062 UP000007801 UP000030765 UP000053825 UP000000803 UP000001819 UP000075883 UP000008792 UP000092461 UP000054359 UP000019118 UP000030742 UP000030746 UP000085678 UP000242188 UP000005408 UP000245300 UP000014760 UP000053286 UP000233160 UP000001555
PRIDE
Gene 3D
ProteinModelPortal
A0A2A4J776
A0A212EXY8
A0A2P8YIB7
K7IVJ9
A0A232EF39
A0A1S3CZ03
+ More
E2C7H7 A0A1B6C963 E2AAJ9 A0A3L8DY01 A0A026WLV0 A0A067QR13 A0A2J7Q9V5 A0A2J7Q9T6 A0A1S4KFX2 E9JDC0 J9K2F5 B0XG38 Q16SL4 A0A1S4FQP4 A0A158NZ16 A0A1Y1N8D2 A0A2R7VUX5 A0A2H8TN53 A0A139WGD2 A0A2S2QQV0 A0A2S2P464 A0A2A3EN76 A0A182FMM8 A0A182SW67 F4WBZ6 U5ER64 A0A0V0G6G6 A0A088A1X0 A0A151WQA1 A0A1L8E333 A0A1L8E3H9 A0A2J7Q9T2 A0A182H4Z9 A0A224XL69 A0A2M4BDU7 A0A195BMF5 A0A336K0T1 A0A2M4BDZ1 A0A2M4BDU6 A0A195FSY4 A0A2M3Z6S2 T1HXA0 A0A2M4ADF2 A0A0K8WM52 A0A151J2P1 A0A0C9QEI3 A0A182IYS0 A0A1J1IVP5 A0A0M3QZ51 A0A1W4VKV0 A0A146L6G1 A0A182W611 A0A1W4V7R0 A0A0A9YFR8 Q7PMP8 B3MS44 A0A0P4W3X9 A0A084VGQ1 A0A0L7QUQ3 Q9VYH2 A0A1W4VK35 Q29IB5 A0A1B6FWX5 A0A1B6L696 A0A182MHM8 A0A2J7Q9V0 B4M726 A0A1B0GL08 A0A087UVX6 A0A1B6J300 A0A1J1IXP3 N6TR11 A0A2P6K3S7 U4UIC4 V4AXE2 A0A1S3IB52 A0A210PSJ7 K1QTT0 S4RPI0 R0KYT2 R7TAR8 A0A087QXS9 A0A2I0U248 A0A2K6GEP2 A0A131XVR8 T1J8E4 B7Q2Y7
E2C7H7 A0A1B6C963 E2AAJ9 A0A3L8DY01 A0A026WLV0 A0A067QR13 A0A2J7Q9V5 A0A2J7Q9T6 A0A1S4KFX2 E9JDC0 J9K2F5 B0XG38 Q16SL4 A0A1S4FQP4 A0A158NZ16 A0A1Y1N8D2 A0A2R7VUX5 A0A2H8TN53 A0A139WGD2 A0A2S2QQV0 A0A2S2P464 A0A2A3EN76 A0A182FMM8 A0A182SW67 F4WBZ6 U5ER64 A0A0V0G6G6 A0A088A1X0 A0A151WQA1 A0A1L8E333 A0A1L8E3H9 A0A2J7Q9T2 A0A182H4Z9 A0A224XL69 A0A2M4BDU7 A0A195BMF5 A0A336K0T1 A0A2M4BDZ1 A0A2M4BDU6 A0A195FSY4 A0A2M3Z6S2 T1HXA0 A0A2M4ADF2 A0A0K8WM52 A0A151J2P1 A0A0C9QEI3 A0A182IYS0 A0A1J1IVP5 A0A0M3QZ51 A0A1W4VKV0 A0A146L6G1 A0A182W611 A0A1W4V7R0 A0A0A9YFR8 Q7PMP8 B3MS44 A0A0P4W3X9 A0A084VGQ1 A0A0L7QUQ3 Q9VYH2 A0A1W4VK35 Q29IB5 A0A1B6FWX5 A0A1B6L696 A0A182MHM8 A0A2J7Q9V0 B4M726 A0A1B0GL08 A0A087UVX6 A0A1B6J300 A0A1J1IXP3 N6TR11 A0A2P6K3S7 U4UIC4 V4AXE2 A0A1S3IB52 A0A210PSJ7 K1QTT0 S4RPI0 R0KYT2 R7TAR8 A0A087QXS9 A0A2I0U248 A0A2K6GEP2 A0A131XVR8 T1J8E4 B7Q2Y7
Ontologies
PATHWAY
GO
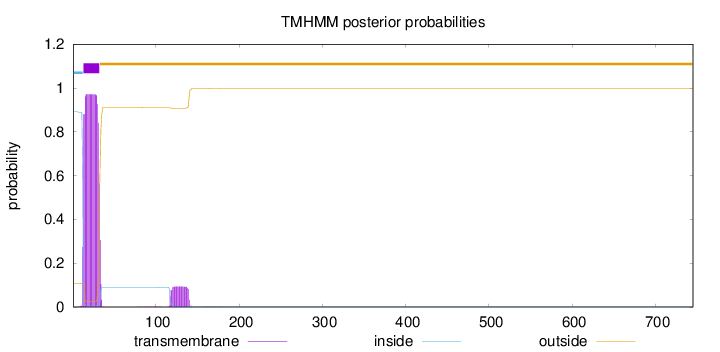
Topology
Subcellular location
Length:
745
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
21.49058
Exp number, first 60 AAs:
19.3716
Total prob of N-in:
0.89164
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 32
outside
33 - 745
Population Genetic Test Statistics
Pi
177.195445
Theta
175.736613
Tajima's D
-1.412697
CLR
14.038324
CSRT
0.0747962601869906
Interpretation
Uncertain
