Gene
KWMTBOMO11812 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013680
Annotation
cytochrome_c_oxidase_polypeptide_IV_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.445
Sequence
CDS
ATGGCCAATTATCTGATGCGCCGGGCGCTCATCAATGCTATCCGTGTTCCTGTCTGCGCCAGGGCTGGATCCACTGGAAACACTGAACTTGCCAAGATTGGTGACCGTGAGTGGGTAGGCTATGGTTTCAACGGCCAGCCCAACTACGTAGACAGGCCTGACTTCCCCCTGCCTGCCATCCGGTTCCGTGAGGACACCCCTGACATTAAGGCTCTCCGTGAAAAAGAAAAGGGTGATTGGCGCAAATTGACCCTCGAAGAAAAGAAAACTCTGTACAGAGCTTCATTCTGTCAGACCTTCGCTGAGTTCCAGGCCCCCACCGGAGAGTGGAAGGGAGTCGTTGGTTGGGCTCTAGTCCTCTCATCTTTGGCTGCTTGGATCTACATGGCCATGAAAGTCTTTGTGTACAGTCCTATTCCTGACTCATTGAGTGAAGAGAGGCAGAAGGCTCAGCTCCAGAGAATGTTGGACCTTAAGGTGAATCCCATCGATGGACTTGCCTCCAAGTGGGACTACGAAAACAACCGCTGGAAGTAA
Protein
MANYLMRRALINAIRVPVCARAGSTGNTELAKIGDREWVGYGFNGQPNYVDRPDFPLPAIRFREDTPDIKALREKEKGDWRKLTLEEKKTLYRASFCQTFAEFQAPTGEWKGVVGWALVLSSLAAWIYMAMKVFVYSPIPDSLSEERQKAQLQRMLDLKVNPIDGLASKWDYENNRWK
Summary
Uniprot
Q1HQ98
A0A2H1W2G2
G8FVP4
A0A3G1T1J1
I4DJE2
E3UKJ2
+ More
A0A212F4W2 S4P8C6 A0A2A4J017 A0A0N0PDN2 A0A194Q5V4 A0A2S1ZDL5 A0A3B0ESC8 A0A1W4WC16 M1R0B9 A0A0P4WTC0 A0A0P5T369 A0A0P5FPC5 A0A0P5FTV3 A0A0P5BPS8 A0A0P5HZR2 A0A0P5WDT0 A0A0P5HL90 A0A0N8B0B2 K9N1H0 A0A0P5J0B6 A0A0P5I2M0 A0A0N8AQV0 A0A0P5G2L3 A0A0P4YZP1 A0A0P4ZT59 A0A0P5X7S2 A0A0P5N5V3 A0A0P5KDI8 A0A0P5PR54 A0A0P5TLY1 A0A0P5EYN5 A0A0P5SIF5 A0A0P5G512 D6WRY0 A0A0P5EFQ9 A0A0P4WNH3 A0A0N7ZM61 A0A0P5GSH5 A0A0P4YLH8 A0A171AJX7 A0A0P5T0N9 A0A224XZL7 A8C9W5 R4WKS7 A0A0N8A4P2 A0A0P4ZLT3 A0A1L8DZS2 A0A0N8CZ60 E9GI31 A0A0P6E0T4 A0A0P5BIK3 A0A0P5T610 A8CWD5 A0A1B6J7K6 A0A0P5PLR2 A0A0P5H589 A0A0P5WL83 A0A0P5Y2V7 A0A1L8DZX0 A0A0T6B5A9 A0A0P5Q6X1 A0A0P4YFB2 R4FL42 A0A0P5H1B7 A0A1B6D2G1 A0A1B6GES7 A0A1Y1JY77 A0A147B9J8 A0A0N8AMJ4 A0A0P5PSF7 A0A069DPY2 U5ERG2 A0A182T6V6 A0A067R5A7 A0A0P4ZZ61 A0A182LRR1 A0A0K8TPZ8 A0A0P4VWS3 A0A2M3ZG18 A0A182YLP4 A0A182PVW9 I4DJY0 A0A2J7RI29 A0A182F3S2 A0A1B6MKP4 W5JUY3 A0A0P6IPI3 A0A182X686 Q7Q8A2 A0A182KWH7 A0A182QSJ2 A0A1S4GZ26 A0A182IBA7 A0A2M4CV40
A0A212F4W2 S4P8C6 A0A2A4J017 A0A0N0PDN2 A0A194Q5V4 A0A2S1ZDL5 A0A3B0ESC8 A0A1W4WC16 M1R0B9 A0A0P4WTC0 A0A0P5T369 A0A0P5FPC5 A0A0P5FTV3 A0A0P5BPS8 A0A0P5HZR2 A0A0P5WDT0 A0A0P5HL90 A0A0N8B0B2 K9N1H0 A0A0P5J0B6 A0A0P5I2M0 A0A0N8AQV0 A0A0P5G2L3 A0A0P4YZP1 A0A0P4ZT59 A0A0P5X7S2 A0A0P5N5V3 A0A0P5KDI8 A0A0P5PR54 A0A0P5TLY1 A0A0P5EYN5 A0A0P5SIF5 A0A0P5G512 D6WRY0 A0A0P5EFQ9 A0A0P4WNH3 A0A0N7ZM61 A0A0P5GSH5 A0A0P4YLH8 A0A171AJX7 A0A0P5T0N9 A0A224XZL7 A8C9W5 R4WKS7 A0A0N8A4P2 A0A0P4ZLT3 A0A1L8DZS2 A0A0N8CZ60 E9GI31 A0A0P6E0T4 A0A0P5BIK3 A0A0P5T610 A8CWD5 A0A1B6J7K6 A0A0P5PLR2 A0A0P5H589 A0A0P5WL83 A0A0P5Y2V7 A0A1L8DZX0 A0A0T6B5A9 A0A0P5Q6X1 A0A0P4YFB2 R4FL42 A0A0P5H1B7 A0A1B6D2G1 A0A1B6GES7 A0A1Y1JY77 A0A147B9J8 A0A0N8AMJ4 A0A0P5PSF7 A0A069DPY2 U5ERG2 A0A182T6V6 A0A067R5A7 A0A0P4ZZ61 A0A182LRR1 A0A0K8TPZ8 A0A0P4VWS3 A0A2M3ZG18 A0A182YLP4 A0A182PVW9 I4DJY0 A0A2J7RI29 A0A182F3S2 A0A1B6MKP4 W5JUY3 A0A0P6IPI3 A0A182X686 Q7Q8A2 A0A182KWH7 A0A182QSJ2 A0A1S4GZ26 A0A182IBA7 A0A2M4CV40
Pubmed
EMBL
BABH01038715
DQ443154
DQ813501
ABF51243.1
ABG67687.1
ODYU01005887
+ More
SOQ47238.1 JN082714 AER27812.1 MG992407 AXY94845.1 AK401410 BAM18032.1 HM449866 ADO33001.1 AGBW02010297 OWR48774.1 GAIX01006001 JAA86559.1 NWSH01004166 PCG65415.1 KQ460141 KPJ17487.1 KQ459460 KPJ00744.1 MF374344 AWK22770.1 RBVL01000247 RKO10148.1 JX847497 JX847498 AGG22607.1 GDRN01015602 JAI67845.1 GDIQ01248857 GDIQ01224116 GDIQ01097752 GDIP01134011 JAK02868.1 JAL69703.1 GDIQ01251858 JAJ99866.1 GDIQ01250408 JAK01317.1 GDIP01185555 GDIQ01248856 JAJ37847.1 JAK02869.1 GDIP01145661 GDIQ01226465 JAJ77741.1 JAK25260.1 GDIP01088220 JAM15495.1 GDIQ01225362 JAK26363.1 GDIQ01225363 JAK26362.1 JQ828862 AFY10818.1 GDIQ01253116 GDIQ01204974 GDIQ01198973 GDIP01141511 JAK46751.1 JAL62203.1 GDIQ01221317 JAK30408.1 GDIQ01251859 JAJ99865.1 GDIQ01250409 JAK01316.1 GDIP01220500 JAJ02902.1 GDIP01212051 JAJ11351.1 GDIP01088219 JAM15496.1 GDIQ01146698 JAL05028.1 GDIQ01185824 JAK65901.1 GDIQ01124902 JAL26824.1 GDIP01129802 GDIQ01053327 LRGB01002864 JAL73912.1 JAN41410.1 KZS05750.1 GDIQ01270329 JAJ81395.1 GDIQ01206425 GDIP01139361 JAK45300.1 JAL64353.1 GDIQ01247158 JAK04567.1 KQ971351 EFA07496.1 GDIQ01270245 JAJ81479.1 GDIP01252984 JAI70417.1 GDIP01231849 JAI91552.1 GDIQ01243261 JAK08464.1 GDIP01227102 JAI96299.1 GEMB01000976 JAS02167.1 GDIP01132814 JAL70900.1 GFTR01002905 JAW13521.1 EU032343 ABV44710.1 AK418220 BAN21435.1 GDIP01182801 JAJ40601.1 GDIP01212898 JAJ10504.1 GFDF01002137 JAV11947.1 GDIP01084567 JAM19148.1 GL732545 EFX80947.1 GDIQ01081264 JAN13473.1 GDIP01184254 JAJ39148.1 GDIP01132813 JAL70901.1 AJWK01020200 AJWK01020201 EU124618 ABV60336.1 GECU01012541 JAS95165.1 GDIQ01126601 JAL25125.1 GDIQ01232868 JAK18857.1 GDIP01084566 JAM19149.1 GDIP01064720 JAM38995.1 GFDF01002110 JAV11974.1 LJIG01009694 KRT82568.1 GDIQ01124903 JAL26823.1 GDIP01231848 JAI91553.1 ACPB03014878 GAHY01001583 JAA75927.1 GDIQ01232869 JAK18856.1 GEDC01017462 JAS19836.1 GECZ01008831 JAS60938.1 GEZM01097356 JAV54259.1 GEIB01000405 JAR87459.1 GDIQ01261107 JAJ90617.1 GDIQ01124904 JAL26822.1 GBGD01003197 JAC85692.1 GANO01003762 JAB56109.1 KK852686 KDR18465.1 GDIP01219150 JAJ04252.1 AXCM01000660 GDAI01001387 JAI16216.1 GDKW01001449 JAI55146.1 GGFM01006639 MBW27390.1 AK401598 BAM18220.1 NEVH01003506 PNF40476.1 GEBQ01003449 JAT36528.1 ADMH02000386 ETN66755.1 GDIQ01002304 JAN92433.1 AAAB01008944 EAA10128.4 AXCN02000628 APCN01000663 GGFL01004883 MBW69061.1
SOQ47238.1 JN082714 AER27812.1 MG992407 AXY94845.1 AK401410 BAM18032.1 HM449866 ADO33001.1 AGBW02010297 OWR48774.1 GAIX01006001 JAA86559.1 NWSH01004166 PCG65415.1 KQ460141 KPJ17487.1 KQ459460 KPJ00744.1 MF374344 AWK22770.1 RBVL01000247 RKO10148.1 JX847497 JX847498 AGG22607.1 GDRN01015602 JAI67845.1 GDIQ01248857 GDIQ01224116 GDIQ01097752 GDIP01134011 JAK02868.1 JAL69703.1 GDIQ01251858 JAJ99866.1 GDIQ01250408 JAK01317.1 GDIP01185555 GDIQ01248856 JAJ37847.1 JAK02869.1 GDIP01145661 GDIQ01226465 JAJ77741.1 JAK25260.1 GDIP01088220 JAM15495.1 GDIQ01225362 JAK26363.1 GDIQ01225363 JAK26362.1 JQ828862 AFY10818.1 GDIQ01253116 GDIQ01204974 GDIQ01198973 GDIP01141511 JAK46751.1 JAL62203.1 GDIQ01221317 JAK30408.1 GDIQ01251859 JAJ99865.1 GDIQ01250409 JAK01316.1 GDIP01220500 JAJ02902.1 GDIP01212051 JAJ11351.1 GDIP01088219 JAM15496.1 GDIQ01146698 JAL05028.1 GDIQ01185824 JAK65901.1 GDIQ01124902 JAL26824.1 GDIP01129802 GDIQ01053327 LRGB01002864 JAL73912.1 JAN41410.1 KZS05750.1 GDIQ01270329 JAJ81395.1 GDIQ01206425 GDIP01139361 JAK45300.1 JAL64353.1 GDIQ01247158 JAK04567.1 KQ971351 EFA07496.1 GDIQ01270245 JAJ81479.1 GDIP01252984 JAI70417.1 GDIP01231849 JAI91552.1 GDIQ01243261 JAK08464.1 GDIP01227102 JAI96299.1 GEMB01000976 JAS02167.1 GDIP01132814 JAL70900.1 GFTR01002905 JAW13521.1 EU032343 ABV44710.1 AK418220 BAN21435.1 GDIP01182801 JAJ40601.1 GDIP01212898 JAJ10504.1 GFDF01002137 JAV11947.1 GDIP01084567 JAM19148.1 GL732545 EFX80947.1 GDIQ01081264 JAN13473.1 GDIP01184254 JAJ39148.1 GDIP01132813 JAL70901.1 AJWK01020200 AJWK01020201 EU124618 ABV60336.1 GECU01012541 JAS95165.1 GDIQ01126601 JAL25125.1 GDIQ01232868 JAK18857.1 GDIP01084566 JAM19149.1 GDIP01064720 JAM38995.1 GFDF01002110 JAV11974.1 LJIG01009694 KRT82568.1 GDIQ01124903 JAL26823.1 GDIP01231848 JAI91553.1 ACPB03014878 GAHY01001583 JAA75927.1 GDIQ01232869 JAK18856.1 GEDC01017462 JAS19836.1 GECZ01008831 JAS60938.1 GEZM01097356 JAV54259.1 GEIB01000405 JAR87459.1 GDIQ01261107 JAJ90617.1 GDIQ01124904 JAL26822.1 GBGD01003197 JAC85692.1 GANO01003762 JAB56109.1 KK852686 KDR18465.1 GDIP01219150 JAJ04252.1 AXCM01000660 GDAI01001387 JAI16216.1 GDKW01001449 JAI55146.1 GGFM01006639 MBW27390.1 AK401598 BAM18220.1 NEVH01003506 PNF40476.1 GEBQ01003449 JAT36528.1 ADMH02000386 ETN66755.1 GDIQ01002304 JAN92433.1 AAAB01008944 EAA10128.4 AXCN02000628 APCN01000663 GGFL01004883 MBW69061.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000053268
UP000276569
+ More
UP000192223 UP000076858 UP000007266 UP000000305 UP000092461 UP000015103 UP000075901 UP000027135 UP000075883 UP000076408 UP000075885 UP000235965 UP000069272 UP000000673 UP000076407 UP000007062 UP000075882 UP000075886 UP000075840
UP000192223 UP000076858 UP000007266 UP000000305 UP000092461 UP000015103 UP000075901 UP000027135 UP000075883 UP000076408 UP000075885 UP000235965 UP000069272 UP000000673 UP000076407 UP000007062 UP000075882 UP000075886 UP000075840
PRIDE
Interpro
Gene 3D
ProteinModelPortal
Q1HQ98
A0A2H1W2G2
G8FVP4
A0A3G1T1J1
I4DJE2
E3UKJ2
+ More
A0A212F4W2 S4P8C6 A0A2A4J017 A0A0N0PDN2 A0A194Q5V4 A0A2S1ZDL5 A0A3B0ESC8 A0A1W4WC16 M1R0B9 A0A0P4WTC0 A0A0P5T369 A0A0P5FPC5 A0A0P5FTV3 A0A0P5BPS8 A0A0P5HZR2 A0A0P5WDT0 A0A0P5HL90 A0A0N8B0B2 K9N1H0 A0A0P5J0B6 A0A0P5I2M0 A0A0N8AQV0 A0A0P5G2L3 A0A0P4YZP1 A0A0P4ZT59 A0A0P5X7S2 A0A0P5N5V3 A0A0P5KDI8 A0A0P5PR54 A0A0P5TLY1 A0A0P5EYN5 A0A0P5SIF5 A0A0P5G512 D6WRY0 A0A0P5EFQ9 A0A0P4WNH3 A0A0N7ZM61 A0A0P5GSH5 A0A0P4YLH8 A0A171AJX7 A0A0P5T0N9 A0A224XZL7 A8C9W5 R4WKS7 A0A0N8A4P2 A0A0P4ZLT3 A0A1L8DZS2 A0A0N8CZ60 E9GI31 A0A0P6E0T4 A0A0P5BIK3 A0A0P5T610 A8CWD5 A0A1B6J7K6 A0A0P5PLR2 A0A0P5H589 A0A0P5WL83 A0A0P5Y2V7 A0A1L8DZX0 A0A0T6B5A9 A0A0P5Q6X1 A0A0P4YFB2 R4FL42 A0A0P5H1B7 A0A1B6D2G1 A0A1B6GES7 A0A1Y1JY77 A0A147B9J8 A0A0N8AMJ4 A0A0P5PSF7 A0A069DPY2 U5ERG2 A0A182T6V6 A0A067R5A7 A0A0P4ZZ61 A0A182LRR1 A0A0K8TPZ8 A0A0P4VWS3 A0A2M3ZG18 A0A182YLP4 A0A182PVW9 I4DJY0 A0A2J7RI29 A0A182F3S2 A0A1B6MKP4 W5JUY3 A0A0P6IPI3 A0A182X686 Q7Q8A2 A0A182KWH7 A0A182QSJ2 A0A1S4GZ26 A0A182IBA7 A0A2M4CV40
A0A212F4W2 S4P8C6 A0A2A4J017 A0A0N0PDN2 A0A194Q5V4 A0A2S1ZDL5 A0A3B0ESC8 A0A1W4WC16 M1R0B9 A0A0P4WTC0 A0A0P5T369 A0A0P5FPC5 A0A0P5FTV3 A0A0P5BPS8 A0A0P5HZR2 A0A0P5WDT0 A0A0P5HL90 A0A0N8B0B2 K9N1H0 A0A0P5J0B6 A0A0P5I2M0 A0A0N8AQV0 A0A0P5G2L3 A0A0P4YZP1 A0A0P4ZT59 A0A0P5X7S2 A0A0P5N5V3 A0A0P5KDI8 A0A0P5PR54 A0A0P5TLY1 A0A0P5EYN5 A0A0P5SIF5 A0A0P5G512 D6WRY0 A0A0P5EFQ9 A0A0P4WNH3 A0A0N7ZM61 A0A0P5GSH5 A0A0P4YLH8 A0A171AJX7 A0A0P5T0N9 A0A224XZL7 A8C9W5 R4WKS7 A0A0N8A4P2 A0A0P4ZLT3 A0A1L8DZS2 A0A0N8CZ60 E9GI31 A0A0P6E0T4 A0A0P5BIK3 A0A0P5T610 A8CWD5 A0A1B6J7K6 A0A0P5PLR2 A0A0P5H589 A0A0P5WL83 A0A0P5Y2V7 A0A1L8DZX0 A0A0T6B5A9 A0A0P5Q6X1 A0A0P4YFB2 R4FL42 A0A0P5H1B7 A0A1B6D2G1 A0A1B6GES7 A0A1Y1JY77 A0A147B9J8 A0A0N8AMJ4 A0A0P5PSF7 A0A069DPY2 U5ERG2 A0A182T6V6 A0A067R5A7 A0A0P4ZZ61 A0A182LRR1 A0A0K8TPZ8 A0A0P4VWS3 A0A2M3ZG18 A0A182YLP4 A0A182PVW9 I4DJY0 A0A2J7RI29 A0A182F3S2 A0A1B6MKP4 W5JUY3 A0A0P6IPI3 A0A182X686 Q7Q8A2 A0A182KWH7 A0A182QSJ2 A0A1S4GZ26 A0A182IBA7 A0A2M4CV40
PDB
5Z62
E-value=7.96961e-29,
Score=312
Ontologies
PATHWAY
GO
PANTHER
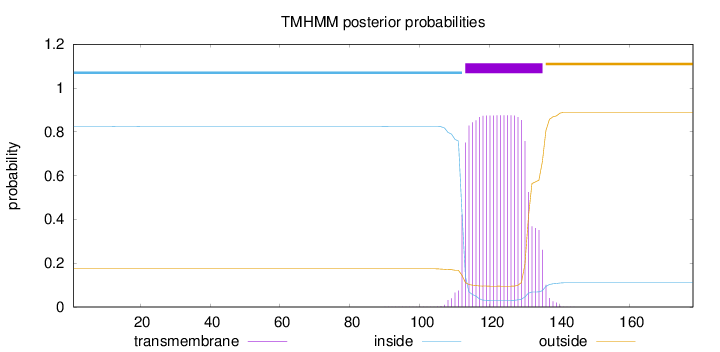
Topology
Length:
178
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
18.08114
Exp number, first 60 AAs:
0.00135
Total prob of N-in:
0.82520
inside
1 - 112
TMhelix
113 - 135
outside
136 - 178
Population Genetic Test Statistics
Pi
267.208198
Theta
186.81115
Tajima's D
0.658117
CLR
0.028032
CSRT
0.562121893905305
Interpretation
Uncertain
