Gene
KWMTBOMO11810
Pre Gene Modal
BGIBMGA013666
Annotation
Cytochrome_c_oxidase_polypeptide_IV_[Operophtera_brumata]
Location in the cell
Mitochondrial Reliability : 2.445
Sequence
CDS
ATGTCGTTACTAGGTCCAAAAAGATTATTTAAATTATCTTTAAATTTAGTTAGAAGTGTTCACAATCGATGTAGGATCGGGAACAGAGACTGGGTCGGTTATGGCGTCAACGGAATGGCCAATTACAAGGACGAAGCGCAATTTCCCTTTCCAGCTGTGAGATTCAAAGAAAACACCAAAGATATCTGGGCCTTGCGCGAGAAAGAGAAGGGAGATTGGAAACTCCTCTGCTGCGAAGAGAAGAAAGCGTTATATCGTGCTTCTTTCTGTCAGACCTTCGCCGAATTTCAGCACTATACAGGAGAATGGAAACTCATCCTGGGATATTTGCTTATCGCTCTGTCTTTTCCCTTTTGGGCAATGATATTCAACCACTATTATGTATATGAACCTCTTCCCGAATCGCTGTCCAAAGAGTCACAAAAGGCTCAATTGAGGAGGATGCTGGAGTTACGCGTCAACCCAATAGACGGATTATCGTCTAAATGGGACTATGATAACGACCGATGGAAGGTAGACTACTTTTATTTTGCTCTAGATCCGTCTAGAAAACTTCGCAACCGTCACCAGAGAAGTCACGGTTAG
Protein
MSLLGPKRLFKLSLNLVRSVHNRCRIGNRDWVGYGVNGMANYKDEAQFPFPAVRFKENTKDIWALREKEKGDWKLLCCEEKKALYRASFCQTFAEFQHYTGEWKLILGYLLIALSFPFWAMIFNHYYVYEPLPESLSKESQKAQLRRMLELRVNPIDGLSSKWDYDNDRWKVDYFYFALDPSRKLRNRHQRSHG
Summary
Uniprot
H9JVV3
A0A0L7L8C7
A0A2H1VRD0
A0A2A4J0K7
A0A2A4J1T7
A0A194QBD8
+ More
A0A212F4W4 A0A0N1IPP8 A0A2H1W2G2 S4P8C6 A0A212F4W2 A0A3G1T1J1 G8FVP4 E3UKJ2 Q1HQ98 A0A2A4J017 U5ERG2 A0A182R414 I4DJE2 A0A3B0ESC8 A0A2S1ZDL5 Q7Q8A2 A0A0K8TPZ8 A0A1S4GZ26 A0A182IBA7 A0A182KWH7 A0A182V8V5 M1R0B9 A0A182X686 A0A182NBB7 A0A182U0F7 A0A182T6V6 A0A0N0PDN2 A0A1W7RAP4 A0A182K4X1 A0A182LRR1 A0A182YLP4 A0A0P4WTC0 A0A182VTH4 A0A194Q5V4 A0A226DRW0 A0A182PVW9 A0A067R5A7 A0A1Y1JY77 A0A182QSJ2 A0A2M4A347 W5JUY3 A0A2M4CUP1 A0A2M4CV40 A0A2J7RI29 A0A2M4C2D4 D6WRY0 T1JMD6 A0A147B9J8 A0A084VMJ5 A0A1W4WC16 A0A182F3S2 A0A2M3ZG18 A0A0L0CDV8 A0A182J1A9 T1E9I0 A0A0T6B5A9 A0A131XVQ0 A0A2R5LH08 A0A2I9LNX1 B0WCJ9 Q4PM46 A0A1D2MHF4 A0A1Z5L5R7 A0A0P6IPI3 B2D274 K9N1H0 T1PFG2 A0A0P4YLH8 A6N9X3 E9GI31 A0A0P5T369 A0A0P5HZR2 A0A0P5BPS8 J3JWR3 A0A0P5GSH5 Q16KF6 A0A0P5N5V3 A0A0P5WDT0 A0A0P5X7S2 A0A0P5TLY1 A0A0P5HL90 A0A0P5J0B6 A0A0N7ZM61 A0A0P5PR54 W8CDW9 A0A0P5EYN5 A0A0P4YZP1 J3JXV3 A0A0P5FPC5 A0A0N8B0B2 A0A0P5I2M0 A0A0N8AQV0 A8C9W5 A0A034WLA2
A0A212F4W4 A0A0N1IPP8 A0A2H1W2G2 S4P8C6 A0A212F4W2 A0A3G1T1J1 G8FVP4 E3UKJ2 Q1HQ98 A0A2A4J017 U5ERG2 A0A182R414 I4DJE2 A0A3B0ESC8 A0A2S1ZDL5 Q7Q8A2 A0A0K8TPZ8 A0A1S4GZ26 A0A182IBA7 A0A182KWH7 A0A182V8V5 M1R0B9 A0A182X686 A0A182NBB7 A0A182U0F7 A0A182T6V6 A0A0N0PDN2 A0A1W7RAP4 A0A182K4X1 A0A182LRR1 A0A182YLP4 A0A0P4WTC0 A0A182VTH4 A0A194Q5V4 A0A226DRW0 A0A182PVW9 A0A067R5A7 A0A1Y1JY77 A0A182QSJ2 A0A2M4A347 W5JUY3 A0A2M4CUP1 A0A2M4CV40 A0A2J7RI29 A0A2M4C2D4 D6WRY0 T1JMD6 A0A147B9J8 A0A084VMJ5 A0A1W4WC16 A0A182F3S2 A0A2M3ZG18 A0A0L0CDV8 A0A182J1A9 T1E9I0 A0A0T6B5A9 A0A131XVQ0 A0A2R5LH08 A0A2I9LNX1 B0WCJ9 Q4PM46 A0A1D2MHF4 A0A1Z5L5R7 A0A0P6IPI3 B2D274 K9N1H0 T1PFG2 A0A0P4YLH8 A6N9X3 E9GI31 A0A0P5T369 A0A0P5HZR2 A0A0P5BPS8 J3JWR3 A0A0P5GSH5 Q16KF6 A0A0P5N5V3 A0A0P5WDT0 A0A0P5X7S2 A0A0P5TLY1 A0A0P5HL90 A0A0P5J0B6 A0A0N7ZM61 A0A0P5PR54 W8CDW9 A0A0P5EYN5 A0A0P4YZP1 J3JXV3 A0A0P5FPC5 A0A0N8B0B2 A0A0P5I2M0 A0A0N8AQV0 A8C9W5 A0A034WLA2
Pubmed
19121390
26227816
26354079
22118469
23622113
22651552
+ More
12364791 26369729 20966253 23390104 25244985 24845553 28004739 20920257 23761445 18362917 19820115 24438588 26108605 29248469 16431279 27289101 28528879 18725333 23831752 25315136 18070662 21292972 22516182 23537049 17510324 24495485 17760985 25348373
12364791 26369729 20966253 23390104 25244985 24845553 28004739 20920257 23761445 18362917 19820115 24438588 26108605 29248469 16431279 27289101 28528879 18725333 23831752 25315136 18070662 21292972 22516182 23537049 17510324 24495485 17760985 25348373
EMBL
BABH01038713
BABH01038714
JTDY01002265
KOB71773.1
ODYU01003960
SOQ43357.1
+ More
NWSH01004166 PCG65409.1 PCG65410.1 KQ459460 KPJ00746.1 AGBW02010297 OWR48777.1 KQ460141 KPJ17485.1 ODYU01005887 SOQ47238.1 GAIX01006001 JAA86559.1 OWR48774.1 MG992407 AXY94845.1 JN082714 AER27812.1 HM449866 ADO33001.1 BABH01038715 DQ443154 DQ813501 ABF51243.1 ABG67687.1 PCG65415.1 GANO01003762 JAB56109.1 AK401410 BAM18032.1 RBVL01000247 RKO10148.1 MF374344 AWK22770.1 AAAB01008944 EAA10128.4 GDAI01001387 JAI16216.1 APCN01000663 JX847497 JX847498 AGG22607.1 KPJ17487.1 GFAH01000171 JAV48218.1 AXCM01000660 GDRN01015602 JAI67845.1 KPJ00744.1 LNIX01000014 OXA46946.1 KK852686 KDR18465.1 GEZM01097356 JAV54259.1 AXCN02000628 GGFK01001868 MBW35189.1 ADMH02000386 ETN66755.1 GGFL01004884 MBW69062.1 GGFL01004883 MBW69061.1 NEVH01003506 PNF40476.1 GGFJ01010346 MBW59487.1 KQ971351 EFA07496.1 JH431429 GEIB01000405 JAR87459.1 ATLV01014599 KE524975 KFB39189.1 GGFM01006639 MBW27390.1 JRES01000523 KNC30430.1 GAMD01001332 JAB00259.1 LJIG01009694 KRT82568.1 GEFM01005511 JAP70285.1 GGLE01004680 MBY08806.1 GFWZ01000092 MBW20082.1 DS231888 EDS43570.1 DQ066281 AAY66918.1 LJIJ01001230 ODM92437.1 GFJQ02004639 JAW02331.1 GDIQ01002304 JAN92433.1 EU574808 ACB70315.1 JQ828862 AFY10818.1 KA647424 AFP62053.1 GDIP01227102 JAI96299.1 EF633939 ABR23456.1 GL732545 EFX80947.1 GDIQ01248857 GDIQ01224116 GDIQ01097752 GDIP01134011 JAK02868.1 JAL69703.1 GDIP01145661 GDIQ01226465 JAJ77741.1 JAK25260.1 GDIP01185555 GDIQ01248856 JAJ37847.1 JAK02869.1 APGK01033474 BT127681 KB740879 KB631953 AEE62643.1 ENN78519.1 ERL87445.1 GDIQ01243261 JAK08464.1 CH477960 EAT34784.1 GDIQ01146698 JAL05028.1 GDIP01088220 JAM15495.1 GDIP01088219 JAM15496.1 GDIP01129802 GDIQ01053327 LRGB01002864 JAL73912.1 JAN41410.1 KZS05750.1 GDIQ01225362 JAK26363.1 GDIQ01253116 GDIQ01204974 GDIQ01198973 GDIP01141511 JAK46751.1 JAL62203.1 GDIP01231849 JAI91552.1 GDIQ01124902 JAL26824.1 GAMC01001216 JAC05340.1 GDIQ01270329 JAJ81395.1 GDIP01220500 JAJ02902.1 BT128075 AEE63036.1 GDIQ01251858 JAJ99866.1 GDIQ01225363 JAK26362.1 GDIQ01221317 JAK30408.1 GDIQ01251859 JAJ99865.1 EU032343 ABV44710.1 GAKP01004027 JAC54925.1
NWSH01004166 PCG65409.1 PCG65410.1 KQ459460 KPJ00746.1 AGBW02010297 OWR48777.1 KQ460141 KPJ17485.1 ODYU01005887 SOQ47238.1 GAIX01006001 JAA86559.1 OWR48774.1 MG992407 AXY94845.1 JN082714 AER27812.1 HM449866 ADO33001.1 BABH01038715 DQ443154 DQ813501 ABF51243.1 ABG67687.1 PCG65415.1 GANO01003762 JAB56109.1 AK401410 BAM18032.1 RBVL01000247 RKO10148.1 MF374344 AWK22770.1 AAAB01008944 EAA10128.4 GDAI01001387 JAI16216.1 APCN01000663 JX847497 JX847498 AGG22607.1 KPJ17487.1 GFAH01000171 JAV48218.1 AXCM01000660 GDRN01015602 JAI67845.1 KPJ00744.1 LNIX01000014 OXA46946.1 KK852686 KDR18465.1 GEZM01097356 JAV54259.1 AXCN02000628 GGFK01001868 MBW35189.1 ADMH02000386 ETN66755.1 GGFL01004884 MBW69062.1 GGFL01004883 MBW69061.1 NEVH01003506 PNF40476.1 GGFJ01010346 MBW59487.1 KQ971351 EFA07496.1 JH431429 GEIB01000405 JAR87459.1 ATLV01014599 KE524975 KFB39189.1 GGFM01006639 MBW27390.1 JRES01000523 KNC30430.1 GAMD01001332 JAB00259.1 LJIG01009694 KRT82568.1 GEFM01005511 JAP70285.1 GGLE01004680 MBY08806.1 GFWZ01000092 MBW20082.1 DS231888 EDS43570.1 DQ066281 AAY66918.1 LJIJ01001230 ODM92437.1 GFJQ02004639 JAW02331.1 GDIQ01002304 JAN92433.1 EU574808 ACB70315.1 JQ828862 AFY10818.1 KA647424 AFP62053.1 GDIP01227102 JAI96299.1 EF633939 ABR23456.1 GL732545 EFX80947.1 GDIQ01248857 GDIQ01224116 GDIQ01097752 GDIP01134011 JAK02868.1 JAL69703.1 GDIP01145661 GDIQ01226465 JAJ77741.1 JAK25260.1 GDIP01185555 GDIQ01248856 JAJ37847.1 JAK02869.1 APGK01033474 BT127681 KB740879 KB631953 AEE62643.1 ENN78519.1 ERL87445.1 GDIQ01243261 JAK08464.1 CH477960 EAT34784.1 GDIQ01146698 JAL05028.1 GDIP01088220 JAM15495.1 GDIP01088219 JAM15496.1 GDIP01129802 GDIQ01053327 LRGB01002864 JAL73912.1 JAN41410.1 KZS05750.1 GDIQ01225362 JAK26363.1 GDIQ01253116 GDIQ01204974 GDIQ01198973 GDIP01141511 JAK46751.1 JAL62203.1 GDIP01231849 JAI91552.1 GDIQ01124902 JAL26824.1 GAMC01001216 JAC05340.1 GDIQ01270329 JAJ81395.1 GDIP01220500 JAJ02902.1 BT128075 AEE63036.1 GDIQ01251858 JAJ99866.1 GDIQ01225363 JAK26362.1 GDIQ01221317 JAK30408.1 GDIQ01251859 JAJ99865.1 EU032343 ABV44710.1 GAKP01004027 JAC54925.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053268
UP000007151
UP000053240
+ More
UP000075900 UP000276569 UP000007062 UP000075840 UP000075882 UP000075903 UP000076407 UP000075884 UP000075902 UP000075901 UP000075881 UP000075883 UP000076408 UP000075920 UP000198287 UP000075885 UP000027135 UP000075886 UP000000673 UP000235965 UP000007266 UP000030765 UP000192223 UP000069272 UP000037069 UP000075880 UP000002320 UP000094527 UP000095301 UP000000305 UP000019118 UP000030742 UP000008820 UP000076858
UP000075900 UP000276569 UP000007062 UP000075840 UP000075882 UP000075903 UP000076407 UP000075884 UP000075902 UP000075901 UP000075881 UP000075883 UP000076408 UP000075920 UP000198287 UP000075885 UP000027135 UP000075886 UP000000673 UP000235965 UP000007266 UP000030765 UP000192223 UP000069272 UP000037069 UP000075880 UP000002320 UP000094527 UP000095301 UP000000305 UP000019118 UP000030742 UP000008820 UP000076858
Interpro
Gene 3D
ProteinModelPortal
H9JVV3
A0A0L7L8C7
A0A2H1VRD0
A0A2A4J0K7
A0A2A4J1T7
A0A194QBD8
+ More
A0A212F4W4 A0A0N1IPP8 A0A2H1W2G2 S4P8C6 A0A212F4W2 A0A3G1T1J1 G8FVP4 E3UKJ2 Q1HQ98 A0A2A4J017 U5ERG2 A0A182R414 I4DJE2 A0A3B0ESC8 A0A2S1ZDL5 Q7Q8A2 A0A0K8TPZ8 A0A1S4GZ26 A0A182IBA7 A0A182KWH7 A0A182V8V5 M1R0B9 A0A182X686 A0A182NBB7 A0A182U0F7 A0A182T6V6 A0A0N0PDN2 A0A1W7RAP4 A0A182K4X1 A0A182LRR1 A0A182YLP4 A0A0P4WTC0 A0A182VTH4 A0A194Q5V4 A0A226DRW0 A0A182PVW9 A0A067R5A7 A0A1Y1JY77 A0A182QSJ2 A0A2M4A347 W5JUY3 A0A2M4CUP1 A0A2M4CV40 A0A2J7RI29 A0A2M4C2D4 D6WRY0 T1JMD6 A0A147B9J8 A0A084VMJ5 A0A1W4WC16 A0A182F3S2 A0A2M3ZG18 A0A0L0CDV8 A0A182J1A9 T1E9I0 A0A0T6B5A9 A0A131XVQ0 A0A2R5LH08 A0A2I9LNX1 B0WCJ9 Q4PM46 A0A1D2MHF4 A0A1Z5L5R7 A0A0P6IPI3 B2D274 K9N1H0 T1PFG2 A0A0P4YLH8 A6N9X3 E9GI31 A0A0P5T369 A0A0P5HZR2 A0A0P5BPS8 J3JWR3 A0A0P5GSH5 Q16KF6 A0A0P5N5V3 A0A0P5WDT0 A0A0P5X7S2 A0A0P5TLY1 A0A0P5HL90 A0A0P5J0B6 A0A0N7ZM61 A0A0P5PR54 W8CDW9 A0A0P5EYN5 A0A0P4YZP1 J3JXV3 A0A0P5FPC5 A0A0N8B0B2 A0A0P5I2M0 A0A0N8AQV0 A8C9W5 A0A034WLA2
A0A212F4W4 A0A0N1IPP8 A0A2H1W2G2 S4P8C6 A0A212F4W2 A0A3G1T1J1 G8FVP4 E3UKJ2 Q1HQ98 A0A2A4J017 U5ERG2 A0A182R414 I4DJE2 A0A3B0ESC8 A0A2S1ZDL5 Q7Q8A2 A0A0K8TPZ8 A0A1S4GZ26 A0A182IBA7 A0A182KWH7 A0A182V8V5 M1R0B9 A0A182X686 A0A182NBB7 A0A182U0F7 A0A182T6V6 A0A0N0PDN2 A0A1W7RAP4 A0A182K4X1 A0A182LRR1 A0A182YLP4 A0A0P4WTC0 A0A182VTH4 A0A194Q5V4 A0A226DRW0 A0A182PVW9 A0A067R5A7 A0A1Y1JY77 A0A182QSJ2 A0A2M4A347 W5JUY3 A0A2M4CUP1 A0A2M4CV40 A0A2J7RI29 A0A2M4C2D4 D6WRY0 T1JMD6 A0A147B9J8 A0A084VMJ5 A0A1W4WC16 A0A182F3S2 A0A2M3ZG18 A0A0L0CDV8 A0A182J1A9 T1E9I0 A0A0T6B5A9 A0A131XVQ0 A0A2R5LH08 A0A2I9LNX1 B0WCJ9 Q4PM46 A0A1D2MHF4 A0A1Z5L5R7 A0A0P6IPI3 B2D274 K9N1H0 T1PFG2 A0A0P4YLH8 A6N9X3 E9GI31 A0A0P5T369 A0A0P5HZR2 A0A0P5BPS8 J3JWR3 A0A0P5GSH5 Q16KF6 A0A0P5N5V3 A0A0P5WDT0 A0A0P5X7S2 A0A0P5TLY1 A0A0P5HL90 A0A0P5J0B6 A0A0N7ZM61 A0A0P5PR54 W8CDW9 A0A0P5EYN5 A0A0P4YZP1 J3JXV3 A0A0P5FPC5 A0A0N8B0B2 A0A0P5I2M0 A0A0N8AQV0 A8C9W5 A0A034WLA2
PDB
2Y69
E-value=2.17975e-23,
Score=266
Ontologies
PATHWAY
GO
PANTHER
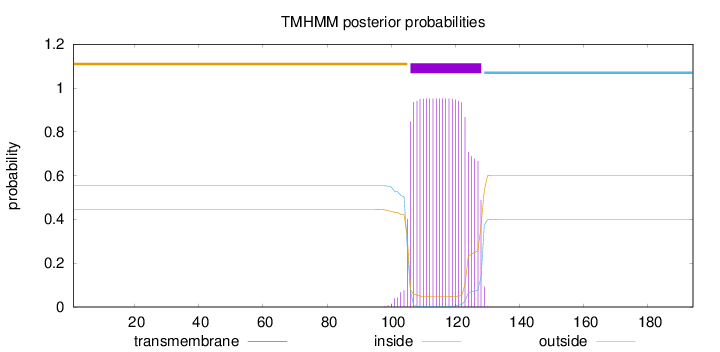
Topology
Length:
194
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.85662
Exp number, first 60 AAs:
0.00063
Total prob of N-in:
0.55391
outside
1 - 105
TMhelix
106 - 128
inside
129 - 194
Population Genetic Test Statistics
Pi
189.889706
Theta
146.684332
Tajima's D
0.766522
CLR
28.187722
CSRT
0.591070446477676
Interpretation
Uncertain
