Gene
KWMTBOMO11809 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013678
Annotation
vacuolar_ATP_synthase_subunit_G_[Bombyx_mori]
Full name
V-type proton ATPase subunit G
+ More
Dolichol-phosphate mannosyltransferase subunit 1
Dolichol-phosphate mannosyltransferase subunit 1
Alternative Name
V-ATPase 13 kDa subunit
Vacuolar proton pump subunit G
Vacuolar proton pump subunit G
Location in the cell
Cytoplasmic Reliability : 1.256 Nuclear Reliability : 1.776
Sequence
CDS
ATGGCGAGTCAGACCCAAGGAATCCAACAGCTTCTAGCTGCTGAAAAACGCGCTGCGGAGAAAGTCAGCGAGGCAAGGAAGCGAAAAGCGAAACGCCTAAAGCAGGCCAAGGAGGAGGCTCAAGATGAAGTTGAAAAGTACAGACAGGAGCGTGAAAGGCAGTTCAAAGAATTTGAAGCCAAGCACATGGGTACCAGGGAAGGTGTTGCGGCCAAGATCGATGCCGAGACCAAAGTTAAGATCGAAGAGATGAACAAAATGGTCCAAACACAGAAGGAGGCGGTCATCAAAGACGTCTTGAATCTGGTGTATGACATCAAACCCGAACTCCACATCAACTACCGCTTAAATTAA
Protein
MASQTQGIQQLLAAEKRAAEKVSEARKRKAKRLKQAKEEAQDEVEKYRQERERQFKEFEAKHMGTREGVAAKIDAETKVKIEEMNKMVQTQKEAVIKDVLNLVYDIKPELHINYRLN
Summary
Description
Catalytic subunit of the peripheral V1 complex of vacuolar ATPase (V-ATPase). V-ATPase is responsible for acidifying a variety of intracellular compartments in eukaryotic cells.
Transfers mannose from GDP-mannose to dolichol monophosphate to form dolichol phosphate mannose (Dol-P-Man) which is the mannosyl donor in pathways leading to N-glycosylation, glycosyl phosphatidylinositol membrane anchoring, and O-mannosylation of proteins.
Transfers mannose from GDP-mannose to dolichol monophosphate to form dolichol phosphate mannose (Dol-P-Man) which is the mannosyl donor in pathways leading to N-glycosylation, glycosyl phosphatidylinositol membrane anchoring, and O-mannosylation of proteins.
Catalytic Activity
dolichyl phosphate + GDP-alpha-D-mannose = dolichyl beta-D-mannosyl phosphate + GDP
Subunit
V-ATPase is a heteromultimeric enzyme composed of a peripheral catalytic V1 complex (components A to H) attached to an integral membrane V0 proton pore complex.
V-ATPase is a heteromultimeric enzyme composed of a peripheral catalytic V1 complex (components A to H) attached to an integral membrane V0 proton pore complex (components: a, c, c', c'' and d).
Component of the dolichol-phosphate mannose (DPM) synthase complex.
V-ATPase is a heteromultimeric enzyme composed of a peripheral catalytic V1 complex (components A to H) attached to an integral membrane V0 proton pore complex (components: a, c, c', c'' and d).
Component of the dolichol-phosphate mannose (DPM) synthase complex.
Similarity
Belongs to the V-ATPase G subunit family.
Belongs to the glycosyltransferase 2 family.
Belongs to the glycosyltransferase 2 family.
Keywords
Cell membrane
Hydrogen ion transport
Ion transport
Membrane
Transport
Feature
chain V-type proton ATPase subunit G
Uniprot
Q2F5J4
S4PD74
Q25532
I4DMP9
A0A0K8TAD3
A0A0N1I9D1
+ More
I4DNL2 A0A3S2L356 A0A1E1W4X6 A0A2H1VRB7 M4MAW1 A0A212F4Z0 D6WPH6 N6UQI0 A0A1L8E0R3 A0A170XKW9 A0A1B6JVS1 A0A1B6G5S6 A0A2S2QVW8 A0A1Y1MM55 A0A069DNI7 Q173D4 Q1W2A2 J3JTX5 U5EYM1 T1DFJ5 A0A023EDV1 A0A232FCE5 K7J8J0 T1E2M7 A0A224Y1G5 A0A336MDG6 A0A1D1ZKP1 A0A2H8TFY3 Q1ZZQ2 B0X4I4 A0A1Q3FGW6 A0A1J1ITM7 E0VSA0 A0A1Q3FH29 A0A0J7KTV3 A0A1B6EDH8 A0A151ITA9 F4WSZ5 A0A195AZ43 A0A158NYI3 C4WWQ7 A0A2R7X2H8 E9IUT5 R4WCX8 A0A195F782 A0A151WQF9 A0A026WMX8 A0A195CWE3 A0A154PB12 U4U5F2 E2A665 B3LXQ4 A0A1S3DCD9 A0A0N0U4C1 A0A1W4W8V4 Q16JK6 E2C0Z3 J9JIR2 A0A0A9YQH2 A0A0F7R132 A0A2A3EEU9 V9IHI2 A0A087ZYW3 B4NFJ4 T1DPT7 A0A182F6W4 W5JSL1 A0A2M3Z5Q7 A0A2M4AG33 A0A2M4C4M3 A0A182FEA8 T1DFY3 A0A0A1XDT7 A0A2M3Z0Y4 A0A2M4A523 W5JCW8 A0A182JMU4 A0A084WJA3 X1X105 A0A0C9QYB4 G9LZB9 A0A0M4EVQ9 B4JYY7 B4K581 B4LYZ4 A0A182LVN6 A0A182RQ45 A0A182WML6 R4G878 T1PP66 A0A3B0JNC8 Q293M1 B4GLX5 A0A1L8EHT7
I4DNL2 A0A3S2L356 A0A1E1W4X6 A0A2H1VRB7 M4MAW1 A0A212F4Z0 D6WPH6 N6UQI0 A0A1L8E0R3 A0A170XKW9 A0A1B6JVS1 A0A1B6G5S6 A0A2S2QVW8 A0A1Y1MM55 A0A069DNI7 Q173D4 Q1W2A2 J3JTX5 U5EYM1 T1DFJ5 A0A023EDV1 A0A232FCE5 K7J8J0 T1E2M7 A0A224Y1G5 A0A336MDG6 A0A1D1ZKP1 A0A2H8TFY3 Q1ZZQ2 B0X4I4 A0A1Q3FGW6 A0A1J1ITM7 E0VSA0 A0A1Q3FH29 A0A0J7KTV3 A0A1B6EDH8 A0A151ITA9 F4WSZ5 A0A195AZ43 A0A158NYI3 C4WWQ7 A0A2R7X2H8 E9IUT5 R4WCX8 A0A195F782 A0A151WQF9 A0A026WMX8 A0A195CWE3 A0A154PB12 U4U5F2 E2A665 B3LXQ4 A0A1S3DCD9 A0A0N0U4C1 A0A1W4W8V4 Q16JK6 E2C0Z3 J9JIR2 A0A0A9YQH2 A0A0F7R132 A0A2A3EEU9 V9IHI2 A0A087ZYW3 B4NFJ4 T1DPT7 A0A182F6W4 W5JSL1 A0A2M3Z5Q7 A0A2M4AG33 A0A2M4C4M3 A0A182FEA8 T1DFY3 A0A0A1XDT7 A0A2M3Z0Y4 A0A2M4A523 W5JCW8 A0A182JMU4 A0A084WJA3 X1X105 A0A0C9QYB4 G9LZB9 A0A0M4EVQ9 B4JYY7 B4K581 B4LYZ4 A0A182LVN6 A0A182RQ45 A0A182WML6 R4G878 T1PP66 A0A3B0JNC8 Q293M1 B4GLX5 A0A1L8EHT7
EC Number
2.4.1.83
Pubmed
19121390
23622113
8626552
22651552
26354079
22118469
+ More
18362917 19820115 23537049 28004739 26334808 17510324 22516182 24330624 24945155 26483478 28648823 20075255 20566863 21719571 21347285 21282665 23691247 24508170 30249741 20798317 17994087 25401762 26823975 25937057 20920257 23761445 25830018 24438588 25348373 25315136 15632085
18362917 19820115 23537049 28004739 26334808 17510324 22516182 24330624 24945155 26483478 28648823 20075255 20566863 21719571 21347285 21282665 23691247 24508170 30249741 20798317 17994087 25401762 26823975 25937057 20920257 23761445 25830018 24438588 25348373 25315136 15632085
EMBL
BABH01038712
BABH01038713
DQ311429
ABD36373.1
GAIX01003776
JAA88784.1
+ More
X92805 AK402567 BAM19189.1 GBRD01003430 JAG62391.1 KQ460141 KPJ17484.1 AK402991 KQ459460 BAM19502.1 KPJ00747.1 RSAL01000195 RVE44574.1 GDQN01009028 JAT82026.1 ODYU01003960 SOQ43360.1 KC127673 AGG56525.1 AGBW02010297 OWR48779.1 KQ971354 EFA06225.1 APGK01020738 KB740234 ENN81002.1 GFDF01001999 JAV12085.1 GEMB01004301 JAR98971.1 GECU01004434 JAT03273.1 GECZ01011981 JAS57788.1 GGMS01012600 MBY81803.1 GEZM01027397 JAV86759.1 GBGD01003529 JAC85360.1 CH477426 EAT41152.1 DQ445525 GEBQ01018984 ABD98763.1 JAT20993.1 BT126685 AEE61647.1 GANO01001918 JAB57953.1 GALA01000562 JAA94290.1 JXUM01038565 GAPW01006121 KQ561170 JAC07477.1 KXJ79476.1 NNAY01000478 OXU28108.1 GALA01000564 JAA94288.1 GFTR01001963 JAW14463.1 UFQS01001008 UFQT01000381 UFQT01001008 SSX08526.1 SSX23761.1 SSX28442.1 GDJX01000468 JAT67468.1 GFXV01000827 MBW12632.1 DQ416024 AK340101 AK340102 AK340103 ABD76371.1 BAH70766.1 DS232342 EDS40401.1 GFDL01008280 JAV26765.1 CVRI01000059 CRL03464.1 DS235748 EEB16256.1 GFDL01008232 JAV26813.1 LBMM01003266 KMQ93731.1 GEDC01001358 JAS35940.1 KQ981029 KYN10197.1 GL888331 EGI62608.1 KQ976701 KYM77305.1 ADTU01003907 AK342213 BAH72327.1 KK856173 PTY25105.1 GL766050 EFZ15654.1 AK417253 BAN20468.1 KQ981768 KYN35929.1 KQ982843 KYQ50043.1 KK107151 QOIP01000003 EZA57293.1 RLU24325.1 KQ977220 KYN04857.1 KQ434864 KZC09099.1 KB632149 ERL89119.1 GL437108 EFN71124.1 CH902617 EDV41711.1 KQ435824 KOX72144.1 CH478004 EAT34456.1 GL451850 EFN78429.1 ABLF02040342 GBHO01010256 GBRD01006996 GDHC01007597 JAG33348.1 JAG58825.1 JAQ11032.1 LC026145 BAR72390.1 KZ288269 PBC30004.1 JR047989 AEY60553.1 CH964251 EDW83061.1 GAMD01003059 JAA98531.1 ADMH02000584 ETN65749.1 GGFM01003095 MBW23846.1 GGFK01006361 MBW39682.1 GGFJ01010787 MBW59928.1 GAMD01002981 JAA98609.1 GBXI01005664 JAD08628.1 GGFM01001429 MBW22180.1 GGFK01002585 MBW35906.1 ADMH02001524 ETN62197.1 ATLV01024003 KE525348 KFB50297.1 ABLF02017433 GBYB01008754 JAG78521.1 JQ218438 GAKP01000834 AEV43310.1 JAC58118.1 CP012526 ALC47467.1 CH916378 EDV98602.1 CH933806 EDW15081.1 CH940650 EDW68097.1 AXCM01002335 ACPB03022079 GAHY01001447 JAA76063.1 KA649786 KA649788 AFP64417.1 OUUW01000008 SPP83737.1 CM000070 EAL29193.1 CH479185 EDW38549.1 GFDG01000558 JAV18241.1
X92805 AK402567 BAM19189.1 GBRD01003430 JAG62391.1 KQ460141 KPJ17484.1 AK402991 KQ459460 BAM19502.1 KPJ00747.1 RSAL01000195 RVE44574.1 GDQN01009028 JAT82026.1 ODYU01003960 SOQ43360.1 KC127673 AGG56525.1 AGBW02010297 OWR48779.1 KQ971354 EFA06225.1 APGK01020738 KB740234 ENN81002.1 GFDF01001999 JAV12085.1 GEMB01004301 JAR98971.1 GECU01004434 JAT03273.1 GECZ01011981 JAS57788.1 GGMS01012600 MBY81803.1 GEZM01027397 JAV86759.1 GBGD01003529 JAC85360.1 CH477426 EAT41152.1 DQ445525 GEBQ01018984 ABD98763.1 JAT20993.1 BT126685 AEE61647.1 GANO01001918 JAB57953.1 GALA01000562 JAA94290.1 JXUM01038565 GAPW01006121 KQ561170 JAC07477.1 KXJ79476.1 NNAY01000478 OXU28108.1 GALA01000564 JAA94288.1 GFTR01001963 JAW14463.1 UFQS01001008 UFQT01000381 UFQT01001008 SSX08526.1 SSX23761.1 SSX28442.1 GDJX01000468 JAT67468.1 GFXV01000827 MBW12632.1 DQ416024 AK340101 AK340102 AK340103 ABD76371.1 BAH70766.1 DS232342 EDS40401.1 GFDL01008280 JAV26765.1 CVRI01000059 CRL03464.1 DS235748 EEB16256.1 GFDL01008232 JAV26813.1 LBMM01003266 KMQ93731.1 GEDC01001358 JAS35940.1 KQ981029 KYN10197.1 GL888331 EGI62608.1 KQ976701 KYM77305.1 ADTU01003907 AK342213 BAH72327.1 KK856173 PTY25105.1 GL766050 EFZ15654.1 AK417253 BAN20468.1 KQ981768 KYN35929.1 KQ982843 KYQ50043.1 KK107151 QOIP01000003 EZA57293.1 RLU24325.1 KQ977220 KYN04857.1 KQ434864 KZC09099.1 KB632149 ERL89119.1 GL437108 EFN71124.1 CH902617 EDV41711.1 KQ435824 KOX72144.1 CH478004 EAT34456.1 GL451850 EFN78429.1 ABLF02040342 GBHO01010256 GBRD01006996 GDHC01007597 JAG33348.1 JAG58825.1 JAQ11032.1 LC026145 BAR72390.1 KZ288269 PBC30004.1 JR047989 AEY60553.1 CH964251 EDW83061.1 GAMD01003059 JAA98531.1 ADMH02000584 ETN65749.1 GGFM01003095 MBW23846.1 GGFK01006361 MBW39682.1 GGFJ01010787 MBW59928.1 GAMD01002981 JAA98609.1 GBXI01005664 JAD08628.1 GGFM01001429 MBW22180.1 GGFK01002585 MBW35906.1 ADMH02001524 ETN62197.1 ATLV01024003 KE525348 KFB50297.1 ABLF02017433 GBYB01008754 JAG78521.1 JQ218438 GAKP01000834 AEV43310.1 JAC58118.1 CP012526 ALC47467.1 CH916378 EDV98602.1 CH933806 EDW15081.1 CH940650 EDW68097.1 AXCM01002335 ACPB03022079 GAHY01001447 JAA76063.1 KA649786 KA649788 AFP64417.1 OUUW01000008 SPP83737.1 CM000070 EAL29193.1 CH479185 EDW38549.1 GFDG01000558 JAV18241.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000283053
UP000007151
UP000007266
+ More
UP000019118 UP000008820 UP000069940 UP000249989 UP000215335 UP000002358 UP000002320 UP000183832 UP000009046 UP000036403 UP000078492 UP000007755 UP000078540 UP000005205 UP000078541 UP000075809 UP000053097 UP000279307 UP000078542 UP000076502 UP000030742 UP000000311 UP000007801 UP000079169 UP000053105 UP000192221 UP000008237 UP000007819 UP000242457 UP000005203 UP000007798 UP000069272 UP000000673 UP000075880 UP000030765 UP000092553 UP000001070 UP000009192 UP000008792 UP000075883 UP000075900 UP000075920 UP000015103 UP000095301 UP000268350 UP000001819 UP000008744
UP000019118 UP000008820 UP000069940 UP000249989 UP000215335 UP000002358 UP000002320 UP000183832 UP000009046 UP000036403 UP000078492 UP000007755 UP000078540 UP000005205 UP000078541 UP000075809 UP000053097 UP000279307 UP000078542 UP000076502 UP000030742 UP000000311 UP000007801 UP000079169 UP000053105 UP000192221 UP000008237 UP000007819 UP000242457 UP000005203 UP000007798 UP000069272 UP000000673 UP000075880 UP000030765 UP000092553 UP000001070 UP000009192 UP000008792 UP000075883 UP000075900 UP000075920 UP000015103 UP000095301 UP000268350 UP000001819 UP000008744
PRIDE
Interpro
SUPFAM
SSF53448
SSF53448
Gene 3D
ProteinModelPortal
Q2F5J4
S4PD74
Q25532
I4DMP9
A0A0K8TAD3
A0A0N1I9D1
+ More
I4DNL2 A0A3S2L356 A0A1E1W4X6 A0A2H1VRB7 M4MAW1 A0A212F4Z0 D6WPH6 N6UQI0 A0A1L8E0R3 A0A170XKW9 A0A1B6JVS1 A0A1B6G5S6 A0A2S2QVW8 A0A1Y1MM55 A0A069DNI7 Q173D4 Q1W2A2 J3JTX5 U5EYM1 T1DFJ5 A0A023EDV1 A0A232FCE5 K7J8J0 T1E2M7 A0A224Y1G5 A0A336MDG6 A0A1D1ZKP1 A0A2H8TFY3 Q1ZZQ2 B0X4I4 A0A1Q3FGW6 A0A1J1ITM7 E0VSA0 A0A1Q3FH29 A0A0J7KTV3 A0A1B6EDH8 A0A151ITA9 F4WSZ5 A0A195AZ43 A0A158NYI3 C4WWQ7 A0A2R7X2H8 E9IUT5 R4WCX8 A0A195F782 A0A151WQF9 A0A026WMX8 A0A195CWE3 A0A154PB12 U4U5F2 E2A665 B3LXQ4 A0A1S3DCD9 A0A0N0U4C1 A0A1W4W8V4 Q16JK6 E2C0Z3 J9JIR2 A0A0A9YQH2 A0A0F7R132 A0A2A3EEU9 V9IHI2 A0A087ZYW3 B4NFJ4 T1DPT7 A0A182F6W4 W5JSL1 A0A2M3Z5Q7 A0A2M4AG33 A0A2M4C4M3 A0A182FEA8 T1DFY3 A0A0A1XDT7 A0A2M3Z0Y4 A0A2M4A523 W5JCW8 A0A182JMU4 A0A084WJA3 X1X105 A0A0C9QYB4 G9LZB9 A0A0M4EVQ9 B4JYY7 B4K581 B4LYZ4 A0A182LVN6 A0A182RQ45 A0A182WML6 R4G878 T1PP66 A0A3B0JNC8 Q293M1 B4GLX5 A0A1L8EHT7
I4DNL2 A0A3S2L356 A0A1E1W4X6 A0A2H1VRB7 M4MAW1 A0A212F4Z0 D6WPH6 N6UQI0 A0A1L8E0R3 A0A170XKW9 A0A1B6JVS1 A0A1B6G5S6 A0A2S2QVW8 A0A1Y1MM55 A0A069DNI7 Q173D4 Q1W2A2 J3JTX5 U5EYM1 T1DFJ5 A0A023EDV1 A0A232FCE5 K7J8J0 T1E2M7 A0A224Y1G5 A0A336MDG6 A0A1D1ZKP1 A0A2H8TFY3 Q1ZZQ2 B0X4I4 A0A1Q3FGW6 A0A1J1ITM7 E0VSA0 A0A1Q3FH29 A0A0J7KTV3 A0A1B6EDH8 A0A151ITA9 F4WSZ5 A0A195AZ43 A0A158NYI3 C4WWQ7 A0A2R7X2H8 E9IUT5 R4WCX8 A0A195F782 A0A151WQF9 A0A026WMX8 A0A195CWE3 A0A154PB12 U4U5F2 E2A665 B3LXQ4 A0A1S3DCD9 A0A0N0U4C1 A0A1W4W8V4 Q16JK6 E2C0Z3 J9JIR2 A0A0A9YQH2 A0A0F7R132 A0A2A3EEU9 V9IHI2 A0A087ZYW3 B4NFJ4 T1DPT7 A0A182F6W4 W5JSL1 A0A2M3Z5Q7 A0A2M4AG33 A0A2M4C4M3 A0A182FEA8 T1DFY3 A0A0A1XDT7 A0A2M3Z0Y4 A0A2M4A523 W5JCW8 A0A182JMU4 A0A084WJA3 X1X105 A0A0C9QYB4 G9LZB9 A0A0M4EVQ9 B4JYY7 B4K581 B4LYZ4 A0A182LVN6 A0A182RQ45 A0A182WML6 R4G878 T1PP66 A0A3B0JNC8 Q293M1 B4GLX5 A0A1L8EHT7
Ontologies
KEGG
PATHWAY
GO
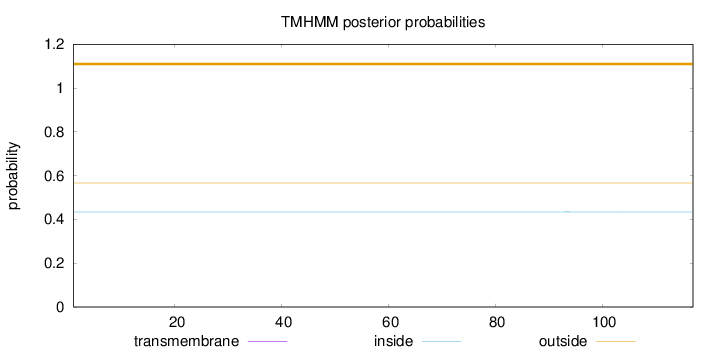
Topology
Subcellular location
Apical cell membrane
Endoplasmic reticulum
Endoplasmic reticulum
Length:
117
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.43422
outside
1 - 117
Population Genetic Test Statistics
Pi
268.288902
Theta
185.816058
Tajima's D
1.244431
CLR
0.263425
CSRT
0.723713814309284
Interpretation
Uncertain
