Gene
KWMTBOMO11806
Pre Gene Modal
BGIBMGA013670
Annotation
PREDICTED:_uncharacterized_protein_LOC101742101_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.426
Sequence
CDS
ATGGAGAAAGCCAAAGCTTACATAAAGAGTTCGTTGGCGACCAAACCTGATTCTCCGACGCTGTTGCGCAAGTCCTCTACTCCGGCCCAGGGTGATGATGTCCGCGTGTGCTTCGTTTGCGGGGGCACCGGCTTCAGTGATCACTACACGATCCGAGTCAAGCCTGATGCCCAGAGCTCGGAGCCTTACTTCCCGTTTTTGGGCGCGCACGCTCCCCCCACCGGCTACAAGGCTGAAGGCGATGACGATGGCACCGTCAAGTGTTGCTGCGTCTGCTACACTTTCCTCCGGCAGCAGTGGGAGCAGTACGACAGAGAGAACAAACCCCACCACCAGAGGTTTTACTGGATGAAGAGATTGGACGGAAAACCTTTTATCGGTGCGGACATGTCCTTCCAAGGTGAATACGCAGCGCAAGTGTTAGGACTCTCGAACGATGCGTCGACTACCCCGGCTCCTCGGGAGGAGCCCCCCGCCGCCTCGCCCCGGCCCACCTCGAGGACTCTATACCCTACTGACCCGGCGCCGCCTGCATCCTCACCTTTCAGGAAAGAAGAGAAGGAAACGAATCATGTCAATAAATTTGTTAATCAGCGGTTTCCCCCCGCGCCCGAGGAGGAGGGCGTGCTGGACCTGCGGGCGCGCGACCCCTCCGTCATATCGGTCGGCTCGCAGCACTCCGGCGCCTCGGAGCCCGCCGGGTACCTGGACGCCGTCAGGTCGACGGTGTCGGTGTACTCGGGCAACTCCAACTCCAGCTCGGACCGCGACATCCTGGACCTGTCGATGCCGGACAAGAACTCGATGACGGAGGTGTGCTACGTCTGCGGCGACGAGTTCAAGCGCGGCACGCTCTACAACCTCAACACCAAGGAGCCCAAGGATAAAACTTTTCAGGCAAAGCAACCCTATTTCCCGATCTTCGGCGAGCAGCACCCCCGGCCGCCGCGCTCCCGGCCCAAGGACGCGCAGGGCACCGTGCGGGCCTGCAGCGCCTGCCACCACCACCTGCTGCAGCAGTGGAACACCTACCAGTTACGAGGGGTACCGGCGAATGAACGCGTCTACACTCTTCGGACCCGTCCCCCCTCCCTACTGCCTCCTTCCCCTCTAAGCCTGGCCAGTTCCGGTGACAAGAGTCTGGCGGCCCACACCCACTCTCCTTCCTTAGAACGTCCCGCCAGCCAGCCGTCAGTGCCTCCGACGACTGCGACCTTCGTGTGCTATGTGTGCGGAGTCAGCACCCCCAGCTCTCAGTTGAGGCTGGTGTACTGCTGCCCCAACCCCGAGAGGGAGCCCTACTACCCGTTTATAACGAGTCTGAAGCCCCACGCTGACGCCAGTCCTATTAGTCCACAGGGTATGGTACAGATATGCGCGGCGTGCTACAAGAGCATACCGCACAAGTACCCGGCGTACGGCGAGAATGGCGAACCCAACCACCATAATAATTCACATACCAATAATATTAGATTTAAGCCGTACGATATAAAGTCGAGCTCGAGTCAGTCGAGCCGGCGGCCCTCCAGCGCGCACTCCGCCAGCCCGCACGGGCAGCTCGTCGCCGGGGAGAACGGCATGGGCCTCTACAGGTGCTACGTGTGCGCGGGGCTGTTCCCGCAGCAGTCGATGGAGTGGCTGTCCACCTCCGCGGAGTACATGAACTCGCACGCCATGCACTTCCCGTGCCTGGGCGCGCGCGGCGGCCGCGTGCTGGCGTGCGCGCGCTGCGTGCGCCACCTCGCCACGCAGTGGGACCTCATGGACGCAGAGAGGGTGCCGCTCGAGCACCGCAGGTACAACATACCGTCGCCCCTACCGTCGAACTCGTCAATGAACGGCGAGCGAGTGATCCCGACGCCGCCGTCCACCACCTCCGACCGGACCGTCGCCAGCAACAGCACGTGCACGTCCATCTACTGCTTCCTGTGCGGGCTGCACTCGGACTTCACGCTGGCGCGCGTGCTGTACGGGCAGCCGCAGGGGAACGCGCCCTACTTCCCGTGCCTGCTCACGCACCAGAGCCACCCCAACGCGGAGCAGCTGCGCGACAACGGCTCCGCGCTCGTCTGCACGTTCTGCTACCACTCGCTGCTCAACCAGTGGCGCCGGTACGAGGCGCTGGGCGGGCAGCCGGCGGAGCGCCGCGTGTACAACACGCACGACTACAACTGCCACCTCTGCGGCGTCAAGACGTACCGGAAGCGGGTGCGCGCGCTGCCCATCAAGGAGTTCCCGTTCCTGGTGCAGCGACGGACTGAAAATTCACTGCTGTTAGAGAACGGTGACTACGCTGTCGTGTGTCTAGATTGTTACGAGAGCTTAAGGACGCAGGCGGCGGACTACGAGCGGCGCGGCGTGCCCGTGGACAAGCGCGAGTACAACTGGCTGCAGCAGCCGCCGCCGCCCGAGGACTCCGCCGACGTCACCATCGCGCGCCTGCCCTCCGGGGACAGGTCCGACAAGCTGATACCGCAATCCCTGGTGCTGCAGCGCGGCTCCAGCAAGCGGCACAGCCCCAAGCAGGCCGCGGCGCTGCTGGCAGACCGCCGCCTCACCCCCAAGCTGGAGAAGGCTTCGGACGCAGTTTTAGTTGATGTCCCCGGCCACTGCATGGTGAGCTCTCGCGTCAAAGCTCGCAGGTTCGGGCCGGGCATGTCCCTCACAGCTAACAATTTTCCTAGAGTTGTAGTTGCAATTACAGCGTATTAG
Protein
MEKAKAYIKSSLATKPDSPTLLRKSSTPAQGDDVRVCFVCGGTGFSDHYTIRVKPDAQSSEPYFPFLGAHAPPTGYKAEGDDDGTVKCCCVCYTFLRQQWEQYDRENKPHHQRFYWMKRLDGKPFIGADMSFQGEYAAQVLGLSNDASTTPAPREEPPAASPRPTSRTLYPTDPAPPASSPFRKEEKETNHVNKFVNQRFPPAPEEEGVLDLRARDPSVISVGSQHSGASEPAGYLDAVRSTVSVYSGNSNSSSDRDILDLSMPDKNSMTEVCYVCGDEFKRGTLYNLNTKEPKDKTFQAKQPYFPIFGEQHPRPPRSRPKDAQGTVRACSACHHHLLQQWNTYQLRGVPANERVYTLRTRPPSLLPPSPLSLASSGDKSLAAHTHSPSLERPASQPSVPPTTATFVCYVCGVSTPSSQLRLVYCCPNPEREPYYPFITSLKPHADASPISPQGMVQICAACYKSIPHKYPAYGENGEPNHHNNSHTNNIRFKPYDIKSSSSQSSRRPSSAHSASPHGQLVAGENGMGLYRCYVCAGLFPQQSMEWLSTSAEYMNSHAMHFPCLGARGGRVLACARCVRHLATQWDLMDAERVPLEHRRYNIPSPLPSNSSMNGERVIPTPPSTTSDRTVASNSTCTSIYCFLCGLHSDFTLARVLYGQPQGNAPYFPCLLTHQSHPNAEQLRDNGSALVCTFCYHSLLNQWRRYEALGGQPAERRVYNTHDYNCHLCGVKTYRKRVRALPIKEFPFLVQRRTENSLLLENGDYAVVCLDCYESLRTQAADYERRGVPVDKREYNWLQQPPPPEDSADVTIARLPSGDRSDKLIPQSLVLQRGSSKRHSPKQAAALLADRRLTPKLEKASDAVLVDVPGHCMVSSRVKARRFGPGMSLTANNFPRVVVAITAY
Summary
Uniprot
A0A194Q5V9
A0A2A4JSE4
A0A0C9RBD4
U4UEC2
U4TVC1
A0A139WEQ2
+ More
A0A139WEX4 A0A1Y1KWY8 A0A1B6JZ63 A0A067QZ92 A0A158NYH9 A0A0L7RDW1 A0A154PB85 A0A026WNN7 A0A3L8DW66 A0A151WHM5 A0A0J7L0T7 A0A151IT97 F4WT00 E2A661 A0A195CXI5 A0A195F636 A0A195AZ36 E9IUT6 E2C0Z9 A0A2C9GPE2 A0A1B6KPR7 A0A2A3EFS5 A0A1W4XSH0 A0A224X5L1 A0A1W4XTE4 A0A1W4XHZ0 A0A0K8UG70 A0A023F143 A0A0K8VWM0 A0A0A1WGR8 E0VHB2 A0A0A1WEI4 A0A0Q9WGP3 A0A0Q9WES4 B4J9C8 A0A0A9XQ57 A0A0J9RIE1 A0A0Q9X9L2 B4KQG9 B4GC07 A0A0R3NML2 Q9U8Q0 Q9W244 A0A0B4KGB8 B4QHI4 A0A1W4VC62 A0A0J9RK20 A0A1W4VQ88 A0A0R1DZ10 B4P5G7 B3NLJ6 A0A0Q5VZE9 A0A3B0JJ53 A0A0R3NSU2 B5E0L7 B4I852 A0A0P9AE80 B3MEE0 T1IA29
A0A139WEX4 A0A1Y1KWY8 A0A1B6JZ63 A0A067QZ92 A0A158NYH9 A0A0L7RDW1 A0A154PB85 A0A026WNN7 A0A3L8DW66 A0A151WHM5 A0A0J7L0T7 A0A151IT97 F4WT00 E2A661 A0A195CXI5 A0A195F636 A0A195AZ36 E9IUT6 E2C0Z9 A0A2C9GPE2 A0A1B6KPR7 A0A2A3EFS5 A0A1W4XSH0 A0A224X5L1 A0A1W4XTE4 A0A1W4XHZ0 A0A0K8UG70 A0A023F143 A0A0K8VWM0 A0A0A1WGR8 E0VHB2 A0A0A1WEI4 A0A0Q9WGP3 A0A0Q9WES4 B4J9C8 A0A0A9XQ57 A0A0J9RIE1 A0A0Q9X9L2 B4KQG9 B4GC07 A0A0R3NML2 Q9U8Q0 Q9W244 A0A0B4KGB8 B4QHI4 A0A1W4VC62 A0A0J9RK20 A0A1W4VQ88 A0A0R1DZ10 B4P5G7 B3NLJ6 A0A0Q5VZE9 A0A3B0JJ53 A0A0R3NSU2 B5E0L7 B4I852 A0A0P9AE80 B3MEE0 T1IA29
Pubmed
EMBL
KQ459460
KPJ00749.1
NWSH01000759
PCG74383.1
GBYB01005630
JAG75397.1
+ More
KB632281 ERL91382.1 KB630834 ERL83893.1 KQ971354 KYB26416.1 KYB26415.1 GEZM01071267 GEZM01071266 JAV65919.1 GECU01003210 JAT04497.1 KK853083 KDR11654.1 ADTU01003883 ADTU01003884 ADTU01003885 ADTU01003886 ADTU01003887 ADTU01003888 ADTU01003889 ADTU01003890 ADTU01003891 ADTU01003892 KQ414613 KOC69004.1 KQ434864 KZC09101.1 KK107151 EZA57291.1 QOIP01000003 RLU24562.1 KQ983114 KYQ47341.1 LBMM01001380 KMQ96412.1 KQ981029 KYN10193.1 GL888331 EGI62613.1 GL437108 EFN71120.1 KQ977220 KYN04854.1 KQ981768 KYN35933.1 KQ976701 KYM77302.1 GL766050 EFZ15671.1 GL451850 EFN78435.1 APCN01000890 GEBQ01026538 JAT13439.1 KZ288269 PBC30006.1 GFTR01008660 JAW07766.1 GDHF01026751 JAI25563.1 GBBI01003704 JAC15008.1 GDHF01009042 JAI43272.1 GBXI01016684 JAC97607.1 DS235165 EEB12768.1 GBXI01017417 JAC96874.1 CH940648 KRF79877.1 KRF79876.1 CH916367 EDW01409.1 GBHO01024385 GBHO01024384 GDHC01007048 JAG19219.1 JAG19220.1 JAQ11581.1 CM002911 KMY95803.1 CH933808 KRG05136.1 EDW10309.1 CH479181 EDW32350.1 CM000071 KRT02139.1 AB032181 BAA86717.1 AE013599 AAF46855.2 AGB93667.1 CM000362 EDX08213.1 KMY95801.1 KMY95804.1 CM000158 KRK00119.1 EDW91798.1 KRK00118.1 CH954179 EDV54912.1 KQS61999.1 OUUW01000001 SPP73256.1 KRT02141.1 EDY69016.1 CH480824 EDW56777.1 CH902619 KPU76290.1 EDV36546.2 ACPB03005093
KB632281 ERL91382.1 KB630834 ERL83893.1 KQ971354 KYB26416.1 KYB26415.1 GEZM01071267 GEZM01071266 JAV65919.1 GECU01003210 JAT04497.1 KK853083 KDR11654.1 ADTU01003883 ADTU01003884 ADTU01003885 ADTU01003886 ADTU01003887 ADTU01003888 ADTU01003889 ADTU01003890 ADTU01003891 ADTU01003892 KQ414613 KOC69004.1 KQ434864 KZC09101.1 KK107151 EZA57291.1 QOIP01000003 RLU24562.1 KQ983114 KYQ47341.1 LBMM01001380 KMQ96412.1 KQ981029 KYN10193.1 GL888331 EGI62613.1 GL437108 EFN71120.1 KQ977220 KYN04854.1 KQ981768 KYN35933.1 KQ976701 KYM77302.1 GL766050 EFZ15671.1 GL451850 EFN78435.1 APCN01000890 GEBQ01026538 JAT13439.1 KZ288269 PBC30006.1 GFTR01008660 JAW07766.1 GDHF01026751 JAI25563.1 GBBI01003704 JAC15008.1 GDHF01009042 JAI43272.1 GBXI01016684 JAC97607.1 DS235165 EEB12768.1 GBXI01017417 JAC96874.1 CH940648 KRF79877.1 KRF79876.1 CH916367 EDW01409.1 GBHO01024385 GBHO01024384 GDHC01007048 JAG19219.1 JAG19220.1 JAQ11581.1 CM002911 KMY95803.1 CH933808 KRG05136.1 EDW10309.1 CH479181 EDW32350.1 CM000071 KRT02139.1 AB032181 BAA86717.1 AE013599 AAF46855.2 AGB93667.1 CM000362 EDX08213.1 KMY95801.1 KMY95804.1 CM000158 KRK00119.1 EDW91798.1 KRK00118.1 CH954179 EDV54912.1 KQS61999.1 OUUW01000001 SPP73256.1 KRT02141.1 EDY69016.1 CH480824 EDW56777.1 CH902619 KPU76290.1 EDV36546.2 ACPB03005093
Proteomes
UP000053268
UP000218220
UP000030742
UP000007266
UP000027135
UP000005205
+ More
UP000053825 UP000076502 UP000053097 UP000279307 UP000075809 UP000036403 UP000078492 UP000007755 UP000000311 UP000078542 UP000078541 UP000078540 UP000008237 UP000075840 UP000242457 UP000192223 UP000009046 UP000008792 UP000001070 UP000009192 UP000008744 UP000001819 UP000000803 UP000000304 UP000192221 UP000002282 UP000008711 UP000268350 UP000001292 UP000007801 UP000015103
UP000053825 UP000076502 UP000053097 UP000279307 UP000075809 UP000036403 UP000078492 UP000007755 UP000000311 UP000078542 UP000078541 UP000078540 UP000008237 UP000075840 UP000242457 UP000192223 UP000009046 UP000008792 UP000001070 UP000009192 UP000008744 UP000001819 UP000000803 UP000000304 UP000192221 UP000002282 UP000008711 UP000268350 UP000001292 UP000007801 UP000015103
Interpro
Gene 3D
ProteinModelPortal
A0A194Q5V9
A0A2A4JSE4
A0A0C9RBD4
U4UEC2
U4TVC1
A0A139WEQ2
+ More
A0A139WEX4 A0A1Y1KWY8 A0A1B6JZ63 A0A067QZ92 A0A158NYH9 A0A0L7RDW1 A0A154PB85 A0A026WNN7 A0A3L8DW66 A0A151WHM5 A0A0J7L0T7 A0A151IT97 F4WT00 E2A661 A0A195CXI5 A0A195F636 A0A195AZ36 E9IUT6 E2C0Z9 A0A2C9GPE2 A0A1B6KPR7 A0A2A3EFS5 A0A1W4XSH0 A0A224X5L1 A0A1W4XTE4 A0A1W4XHZ0 A0A0K8UG70 A0A023F143 A0A0K8VWM0 A0A0A1WGR8 E0VHB2 A0A0A1WEI4 A0A0Q9WGP3 A0A0Q9WES4 B4J9C8 A0A0A9XQ57 A0A0J9RIE1 A0A0Q9X9L2 B4KQG9 B4GC07 A0A0R3NML2 Q9U8Q0 Q9W244 A0A0B4KGB8 B4QHI4 A0A1W4VC62 A0A0J9RK20 A0A1W4VQ88 A0A0R1DZ10 B4P5G7 B3NLJ6 A0A0Q5VZE9 A0A3B0JJ53 A0A0R3NSU2 B5E0L7 B4I852 A0A0P9AE80 B3MEE0 T1IA29
A0A139WEX4 A0A1Y1KWY8 A0A1B6JZ63 A0A067QZ92 A0A158NYH9 A0A0L7RDW1 A0A154PB85 A0A026WNN7 A0A3L8DW66 A0A151WHM5 A0A0J7L0T7 A0A151IT97 F4WT00 E2A661 A0A195CXI5 A0A195F636 A0A195AZ36 E9IUT6 E2C0Z9 A0A2C9GPE2 A0A1B6KPR7 A0A2A3EFS5 A0A1W4XSH0 A0A224X5L1 A0A1W4XTE4 A0A1W4XHZ0 A0A0K8UG70 A0A023F143 A0A0K8VWM0 A0A0A1WGR8 E0VHB2 A0A0A1WEI4 A0A0Q9WGP3 A0A0Q9WES4 B4J9C8 A0A0A9XQ57 A0A0J9RIE1 A0A0Q9X9L2 B4KQG9 B4GC07 A0A0R3NML2 Q9U8Q0 Q9W244 A0A0B4KGB8 B4QHI4 A0A1W4VC62 A0A0J9RK20 A0A1W4VQ88 A0A0R1DZ10 B4P5G7 B3NLJ6 A0A0Q5VZE9 A0A3B0JJ53 A0A0R3NSU2 B5E0L7 B4I852 A0A0P9AE80 B3MEE0 T1IA29
Ontologies
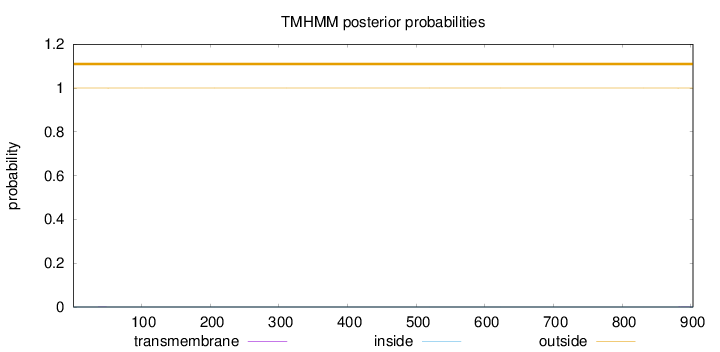
Topology
Length:
903
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00834000000000001
Exp number, first 60 AAs:
0.00215
Total prob of N-in:
0.00022
outside
1 - 903
Population Genetic Test Statistics
Pi
233.58991
Theta
173.516295
Tajima's D
0.781177
CLR
0.612308
CSRT
0.60101994900255
Interpretation
Uncertain
