Gene
KWMTBOMO11793
Pre Gene Modal
BGIBMGA013651
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-like_[Amyelois_transitella]
Full name
Facilitated trehalose transporter Tret1-2 homolog
+ More
Facilitated trehalose transporter Tret1-1
Facilitated trehalose transporter Tret1-1
Location in the cell
PlasmaMembrane Reliability : 4.57
Sequence
CDS
ATGACGTTCGCGTCCGCCATCTTATCATCGTTCGGCGTGACTGTCAGTCCGGAGCTGCAGGCGTTGAGCTTCCCTGCCGTGATGCTGCTCGCCTCCCTGCTGTCACTCTTCTACGTCGAAAGGCTGGGTAGGAAGGTAAGGCCGCCGACCCTGCAGATTAAGAGTGTCCTCACACCACCCGTATGTCAGCACTACAGCGGTAACCGGGATACAAAACTGCTCGGCTCGCGTACGAACATGTCCGAGCTTGAATACACACCGGCGTTTTTGGTAACGGCGGGCTCGCTGGTCTGCCTGGCGACCACCCTGCTCGCCCAGCAGCACGGCTGGGCGGCGCCGGCGTGGCTGCCCGCGGCCGCCATGGTGCTGTCGCTTGCGTCCGGAGTCTCGCCTGTGCCCTATATCATTATGTCTGAAATGTTTCATTTCCAGATTCGCGCCAAGGTGTTGGCTTGCGTGGTGACGCACGCGTGGTTCATGTCGTTCACCCAGCTGGCGGCGTTCGGCCCCTTGTCGGCCGCGCTGGGACTGCACTCCACGTTCTTCGCGTTCGCCGCGGTGAACCTCTGCGGGGCCCTTGTGTCTCTGACGCTCTTGCCCGAAACGAAAGGAAAGACTCTGGAACAAATCGAAATTTCGTTACGAGGGAAACATTTCTTTAGATAA
Protein
MTFASAILSSFGVTVSPELQALSFPAVMLLASLLSLFYVERLGRKVRPPTLQIKSVLTPPVCQHYSGNRDTKLLGSRTNMSELEYTPAFLVTAGSLVCLATTLLAQQHGWAAPAWLPAAAMVLSLASGVSPVPYIIMSEMFHFQIRAKVLACVVTHAWFMSFTQLAAFGPLSAALGLHSTFFAFAAVNLCGALVSLTLLPETKGKTLEQIEISLRGKHFFR
Summary
Description
Fails to transport trehalose.
Low-capacity facilitative transporter for trehalose. Does not transport maltose, sucrose or lactose. Mediates the bidirectional transfer of trehalose. Responsible for the transport of trehalose synthesized in the fat body and the incorporation of trehalose into other tissues that require a carbon source, thereby regulating trehalose levels in the hemolymph.
Low-capacity facilitative transporter for trehalose. Does not transport maltose, sucrose or lactose. Mediates the bidirectional transfer of trehalose. Responsible for the transport of trehalose synthesized in the fat body and the incorporation of trehalose into other tissues that require a carbon source, thereby regulating trehalose levels in the hemolymph.
Biophysicochemical Properties
10.94 mM for trehalose
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family. Trehalose transporter subfamily.
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family. Trehalose transporter subfamily.
Keywords
Cell membrane
Complete proteome
Glycoprotein
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Alternative splicing
Phosphoprotein
Sugar transport
Transport
Feature
chain Facilitated trehalose transporter Tret1-2 homolog
splice variant In isoform B.
splice variant In isoform B.
Uniprot
H9JVT8
A0A2A4J9N1
A0A2H1VLY6
A0A2A4JNL8
A0A194QBF3
A0A194Q5D6
+ More
A0A0N1IJR5 A0A0L7L525 H9JW16 A0A212EZE7 A0A2H1X046 A0A212F0M2 A0A194Q5F7 A0A0N1IIG7 D4AHW6 A0A2A4J8P9 A0A2W1BRA6 A5Y0C3 A0A3S2TMD3 H9J4J6 A0A3S2TIH7 H9JFD4 A0A2H1WGI4 A0A2H1WM77 A0A2A4JKE4 A0A2A4JPP7 A0A2W1BU14 A0A2H1VB69 Q8MKK4 K7IP31 A0A232F7N7 A0A0T6B7J6 A1Z8N1-2 A0A1W4WZ04 A0A0J9RBH4 B4QBN3 A0A0N0PCI3 A0A212F416 A0A2P8ZLZ8 A0A194Q5B9 A0A0J9RAI5 B4HNS1 H9JVU3 A0A212FAG2 A0A194PLS6 A0A1W4WPV3 A0A0L7RBJ5 A0A3S2NG77 A0A0N1IGQ6
A0A0N1IJR5 A0A0L7L525 H9JW16 A0A212EZE7 A0A2H1X046 A0A212F0M2 A0A194Q5F7 A0A0N1IIG7 D4AHW6 A0A2A4J8P9 A0A2W1BRA6 A5Y0C3 A0A3S2TMD3 H9J4J6 A0A3S2TIH7 H9JFD4 A0A2H1WGI4 A0A2H1WM77 A0A2A4JKE4 A0A2A4JPP7 A0A2W1BU14 A0A2H1VB69 Q8MKK4 K7IP31 A0A232F7N7 A0A0T6B7J6 A1Z8N1-2 A0A1W4WZ04 A0A0J9RBH4 B4QBN3 A0A0N0PCI3 A0A212F416 A0A2P8ZLZ8 A0A194Q5B9 A0A0J9RAI5 B4HNS1 H9JVU3 A0A212FAG2 A0A194PLS6 A0A1W4WPV3 A0A0L7RBJ5 A0A3S2NG77 A0A0N1IGQ6
Pubmed
EMBL
BABH01040665
BABH01040666
BABH01040667
NWSH01002244
PCG68837.1
ODYU01003284
+ More
SOQ41841.1 NWSH01001033 PCG73010.1 KQ459460 KPJ00761.1 KPJ00762.1 KQ459838 KPJ19754.1 JTDY01002946 KOB70431.1 BABH01036553 AGBW02011317 OWR46831.1 ODYU01012350 SOQ58622.1 AGBW02011050 OWR47308.1 KPJ00778.1 KPJ19742.1 AB549994 BAI83415.1 NWSH01002620 PCG67830.1 KZ149924 PZC77632.1 EF028238 ABM01870.1 RSAL01000053 RVE50024.1 BABH01021836 RSAL01000114 RVE46930.1 BABH01038299 ODYU01008186 SOQ51574.1 ODYU01009532 SOQ54032.1 NWSH01001270 PCG71903.1 NWSH01000916 PCG73554.1 KZ149936 PZC77145.1 ODYU01001615 SOQ38089.1 AB369551 AE013599 AY122079 AAM52591.1 AAM68715.1 BAF96745.1 NNAY01000793 OXU26469.1 LJIG01009574 KRT82807.1 AB369552 AB369553 BT003466 BT010286 AAF58632.2 AAQ23604.1 BAF96746.1 CM002911 KMY93024.1 CM000362 EDX06650.1 KQ460569 KPJ13961.1 AGBW02010450 OWR48486.1 PYGN01000019 PSN57528.1 KPJ00738.1 KMY93022.1 CH480816 EDW47436.1 BABH01040655 AGBW02009482 OWR50710.1 KQ459601 KPI93689.1 KQ414617 KOC68302.1 RSAL01000118 RVE46763.1 KQ459910 KPJ19331.1
SOQ41841.1 NWSH01001033 PCG73010.1 KQ459460 KPJ00761.1 KPJ00762.1 KQ459838 KPJ19754.1 JTDY01002946 KOB70431.1 BABH01036553 AGBW02011317 OWR46831.1 ODYU01012350 SOQ58622.1 AGBW02011050 OWR47308.1 KPJ00778.1 KPJ19742.1 AB549994 BAI83415.1 NWSH01002620 PCG67830.1 KZ149924 PZC77632.1 EF028238 ABM01870.1 RSAL01000053 RVE50024.1 BABH01021836 RSAL01000114 RVE46930.1 BABH01038299 ODYU01008186 SOQ51574.1 ODYU01009532 SOQ54032.1 NWSH01001270 PCG71903.1 NWSH01000916 PCG73554.1 KZ149936 PZC77145.1 ODYU01001615 SOQ38089.1 AB369551 AE013599 AY122079 AAM52591.1 AAM68715.1 BAF96745.1 NNAY01000793 OXU26469.1 LJIG01009574 KRT82807.1 AB369552 AB369553 BT003466 BT010286 AAF58632.2 AAQ23604.1 BAF96746.1 CM002911 KMY93024.1 CM000362 EDX06650.1 KQ460569 KPJ13961.1 AGBW02010450 OWR48486.1 PYGN01000019 PSN57528.1 KPJ00738.1 KMY93022.1 CH480816 EDW47436.1 BABH01040655 AGBW02009482 OWR50710.1 KQ459601 KPI93689.1 KQ414617 KOC68302.1 RSAL01000118 RVE46763.1 KQ459910 KPJ19331.1
Proteomes
Pfam
PF00083 Sugar_tr
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JVT8
A0A2A4J9N1
A0A2H1VLY6
A0A2A4JNL8
A0A194QBF3
A0A194Q5D6
+ More
A0A0N1IJR5 A0A0L7L525 H9JW16 A0A212EZE7 A0A2H1X046 A0A212F0M2 A0A194Q5F7 A0A0N1IIG7 D4AHW6 A0A2A4J8P9 A0A2W1BRA6 A5Y0C3 A0A3S2TMD3 H9J4J6 A0A3S2TIH7 H9JFD4 A0A2H1WGI4 A0A2H1WM77 A0A2A4JKE4 A0A2A4JPP7 A0A2W1BU14 A0A2H1VB69 Q8MKK4 K7IP31 A0A232F7N7 A0A0T6B7J6 A1Z8N1-2 A0A1W4WZ04 A0A0J9RBH4 B4QBN3 A0A0N0PCI3 A0A212F416 A0A2P8ZLZ8 A0A194Q5B9 A0A0J9RAI5 B4HNS1 H9JVU3 A0A212FAG2 A0A194PLS6 A0A1W4WPV3 A0A0L7RBJ5 A0A3S2NG77 A0A0N1IGQ6
A0A0N1IJR5 A0A0L7L525 H9JW16 A0A212EZE7 A0A2H1X046 A0A212F0M2 A0A194Q5F7 A0A0N1IIG7 D4AHW6 A0A2A4J8P9 A0A2W1BRA6 A5Y0C3 A0A3S2TMD3 H9J4J6 A0A3S2TIH7 H9JFD4 A0A2H1WGI4 A0A2H1WM77 A0A2A4JKE4 A0A2A4JPP7 A0A2W1BU14 A0A2H1VB69 Q8MKK4 K7IP31 A0A232F7N7 A0A0T6B7J6 A1Z8N1-2 A0A1W4WZ04 A0A0J9RBH4 B4QBN3 A0A0N0PCI3 A0A212F416 A0A2P8ZLZ8 A0A194Q5B9 A0A0J9RAI5 B4HNS1 H9JVU3 A0A212FAG2 A0A194PLS6 A0A1W4WPV3 A0A0L7RBJ5 A0A3S2NG77 A0A0N1IGQ6
PDB
6N3I
E-value=0.000153497,
Score=104
Ontologies
GO
Topology
Subcellular location
Cell membrane
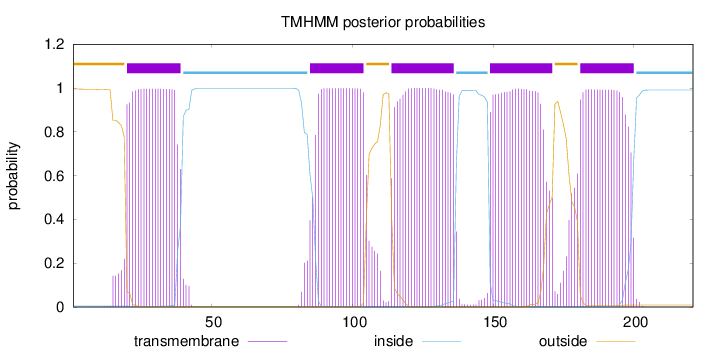
Length:
221
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
106.42864
Exp number, first 60 AAs:
20.3189
Total prob of N-in:
0.00469
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 39
inside
40 - 84
TMhelix
85 - 104
outside
105 - 113
TMhelix
114 - 136
inside
137 - 148
TMhelix
149 - 171
outside
172 - 180
TMhelix
181 - 200
inside
201 - 221
Population Genetic Test Statistics
Pi
292.206904
Theta
198.416057
Tajima's D
1.65777
CLR
0.15449
CSRT
0.821908904554772
Interpretation
Uncertain
