Gene
KWMTBOMO11792
Pre Gene Modal
BGIBMGA013652
Annotation
PREDICTED:_monosaccharide-sensing_protein_3-like_[Plutella_xylostella]
Location in the cell
PlasmaMembrane Reliability : 4.975
Sequence
CDS
ATGAAGGGAAAACTCATACAATATGCGTGCACGGTCGTTTTGACAATACCCTGCTTCCAAGCAGCCTTCACCTTCGTCTGGCCATCATATGCCATACCCGCGCTCCAGAGTGACCGCTCGCCTTTGGGGTACAAGATATCGGAGAGAGAGGCCGGTCTCATCAGCTCTTGCACCATGCTGAGTCCGATGATAGTGACCATGTTCGCCGGAACGCTCGCCGATGTATTCGGAAGAAAGAAGATTTCAGTGGCGTGTGGCTTCATCATTCTTTCATCCTGGATACTAGTGCTCACGGCGAAGTCAGCAAGCCAAATCCTCATAGCTCGACTGATCTGCGGACTGGCGTTCGGTTGCAACATCATTGTGGTCATCATGTACATCGGCGAGCTGTGCCAGACCTCGACGCGCGCCACCGCCACCGTCATCATTACCTTCTTGTTTAGCATTGGAGCCCTGGCATCTTACTCGCTGGGCTGGGTCTGTTCCTATGAGACCGTGTGCTATTTCTGCATCACATTGTCCGTGATCTACATTATATTCATGTTCGCCCTGGTTAAGGAGTCTCCGGTGTTTTTGGTCGGCAAAGGACGGGAAAAGGAAGCGTTACATTGCTTAGCGTTTTATCGCGGCGCGTCAGTCGTTACGGAACACGTCTTGGGCGAGCTCTCGTACATGCGCAGTCAACACAACCAGAACAAACAAAATCATCCCATTGAAGAAAATGTACCTCAATACGAAACCGAGAAGTTAACAGAAGAAAAAACAAGTTTGCCGCAAGAAGAAACAAGTGCATGGAAATTGCTTTTTGCATCAACATCATCACGACGAGCCCTCATCAGCATGGTGTCGATCATGGTCCTGACGGTCTATATGGGGATGGTCAGCGTTCAAGTGTACGCCGCACAACTATTCACGAAGATGGCTCCGAATATTTCACCAGATCTCTGCTCAGTGATTTTAGCTCTCGTCTTGGTGGCCTCCAGCTGTGTCGGCACATTCACGATGGATTTGTTTAAAAGAAGGAGTCTAATGATCTGCTCGACGCTCATGTCGTCCTGGTGCATGGCCGTGCTTGGCAGTCTGCTGCGCTGGTCGTGGGCTTCGGACTGGGCGGTGACGCTCGTCGTCGTTATTTATTGTTTCAGCTATCAAGTGGGACCAAATAGCCTACCGTTCGTGTTGACATCGGAAACTTTCGGCCCCCAGACTAGAGGTGTGGCATCTCAAGTGGTAGTGACATCGATGTTCTTATCAAATTTTCTACTGTTGATACTTTTCGGACCTGTCGAAGATCAGATAGGCTTAGACGGAACATTCTTCCTCTTTTCCATCATTGGATTTGTCACGGTGATATTTACATATTTCACAATGAAGGAAACCAAAGGGATACCGTTGGACGAAATTCAAAAAATGTATGAGAAAGGATTTTTGTATCGTGAAAAATAA
Protein
MKGKLIQYACTVVLTIPCFQAAFTFVWPSYAIPALQSDRSPLGYKISEREAGLISSCTMLSPMIVTMFAGTLADVFGRKKISVACGFIILSSWILVLTAKSASQILIARLICGLAFGCNIIVVIMYIGELCQTSTRATATVIITFLFSIGALASYSLGWVCSYETVCYFCITLSVIYIIFMFALVKESPVFLVGKGREKEALHCLAFYRGASVVTEHVLGELSYMRSQHNQNKQNHPIEENVPQYETEKLTEEKTSLPQEETSAWKLLFASTSSRRALISMVSIMVLTVYMGMVSVQVYAAQLFTKMAPNISPDLCSVILALVLVASSCVGTFTMDLFKRRSLMICSTLMSSWCMAVLGSLLRWSWASDWAVTLVVVIYCFSYQVGPNSLPFVLTSETFGPQTRGVASQVVVTSMFLSNFLLLILFGPVEDQIGLDGTFFLFSIIGFVTVIFTYFTMKETKGIPLDEIQKMYEKGFLYREK
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Belongs to the TRAFAC class myosin-kinesin ATPase superfamily. Myosin family.
Uniprot
H9JVT9
A0A2W1BV94
A0A2A4K0E5
A0A2H1WVK7
A0A0N1IQK5
A0A0N1PKK0
+ More
A0A2A4JUG4 A0A3S2LR68 H9JVU1 A0A194Q6Z4 A0A2W1BZ84 A0A194QBG3 A0A2W1BTX0 A0A2H1VA59 A0A2A4JW98 A0A194Q5E7 A0A194Q5E1 A0A3S2LEN2 A0A2H1X1H2 A0A2A4JW00 A0A0N0PEG4 A0A212F787 A0A0N0PF14 A0A2A4JV69 A0A2W1BPY0 A0A3S2M6I8 H9JWY9 A0A2H1WYH9 A0A2W1BTZ9 A0A2A4JYQ5 A0A2W1BWR1 A0A2J7PIQ0 A0A195AT95 F4W7K1 A0A3L8D924 A0A026X2K8 A0A158NPP3 A0A1B6FS48 A0A1B6MT94 A0A026WQ15 A0A212F6E9 A0A0K8SCV7 A0A146LDU6 A0A0K8SB18 A0A0C9RCE9 A0A0A9YZV0 A0A067QP79 A0A1Y1KQ78 T1DE67 A0A212ERK6 A0A2R7W0J8 A0A1B6M0Z3 A0A1B6FYZ7 A0A1B6MPN8 A0A1Q3FNQ8 A0A1I8NJI3 D4AHW6 T1PBE5 A0A0L0CG94 A0A1B6LM42 A0A182GVZ2 A0A2H1X046 A0A0A8J8K5 A0A2M4A0I2 A0A1W4XL19 A0A0A8J7X6 A0A2M4A0H9 A0A182MM04 A0A182QT48 W5J543 A0A067QZV9 A0A182TSI6 A0A1Q3FQA6 A0A1Q3FQG8 A0A088ABY6 A0A1Q3FQD3 A0A1Q3FQH2 A0A1B6ECS3 A0A182S579 A0A026WS40 A0A1B6FZZ0 T1DG93 A0A1W4XAC6 A0A0N1PEX2 A0A1B6IP97 Q16SU4 A0A182KX77 A0NBZ0 A0A1I8P6M8 A0A067R7D8 A0A182VIR9 A0A182VS58 A0A1B0B100 A0A1Y9J057 A0A0P4VMZ3 A0A2P8XJ31
A0A2A4JUG4 A0A3S2LR68 H9JVU1 A0A194Q6Z4 A0A2W1BZ84 A0A194QBG3 A0A2W1BTX0 A0A2H1VA59 A0A2A4JW98 A0A194Q5E7 A0A194Q5E1 A0A3S2LEN2 A0A2H1X1H2 A0A2A4JW00 A0A0N0PEG4 A0A212F787 A0A0N0PF14 A0A2A4JV69 A0A2W1BPY0 A0A3S2M6I8 H9JWY9 A0A2H1WYH9 A0A2W1BTZ9 A0A2A4JYQ5 A0A2W1BWR1 A0A2J7PIQ0 A0A195AT95 F4W7K1 A0A3L8D924 A0A026X2K8 A0A158NPP3 A0A1B6FS48 A0A1B6MT94 A0A026WQ15 A0A212F6E9 A0A0K8SCV7 A0A146LDU6 A0A0K8SB18 A0A0C9RCE9 A0A0A9YZV0 A0A067QP79 A0A1Y1KQ78 T1DE67 A0A212ERK6 A0A2R7W0J8 A0A1B6M0Z3 A0A1B6FYZ7 A0A1B6MPN8 A0A1Q3FNQ8 A0A1I8NJI3 D4AHW6 T1PBE5 A0A0L0CG94 A0A1B6LM42 A0A182GVZ2 A0A2H1X046 A0A0A8J8K5 A0A2M4A0I2 A0A1W4XL19 A0A0A8J7X6 A0A2M4A0H9 A0A182MM04 A0A182QT48 W5J543 A0A067QZV9 A0A182TSI6 A0A1Q3FQA6 A0A1Q3FQG8 A0A088ABY6 A0A1Q3FQD3 A0A1Q3FQH2 A0A1B6ECS3 A0A182S579 A0A026WS40 A0A1B6FZZ0 T1DG93 A0A1W4XAC6 A0A0N1PEX2 A0A1B6IP97 Q16SU4 A0A182KX77 A0NBZ0 A0A1I8P6M8 A0A067R7D8 A0A182VIR9 A0A182VS58 A0A1B0B100 A0A1Y9J057 A0A0P4VMZ3 A0A2P8XJ31
Pubmed
EMBL
BABH01040661
BABH01040662
BABH01040663
KZ149937
PZC77117.1
NWSH01000350
+ More
PCG77150.1 ODYU01011381 SOQ57088.1 LADI01012895 KPJ20745.1 LADJ01037929 KPJ21328.1 NWSH01000558 PCG75665.1 RSAL01000025 RVE52106.1 BABH01040660 KQ459460 KPJ00770.1 PZC77103.1 KPJ00771.1 PZC77105.1 ODYU01001454 SOQ37676.1 PCG75662.1 KPJ00768.1 KPJ00767.1 RVE52115.1 ODYU01012723 SOQ59169.1 PCG75663.1 KQ459838 KPJ19748.1 AGBW02009924 OWR49569.1 KPJ20744.1 PCG75666.1 PZC77102.1 RVE52104.1 BABH01043549 ODYU01011969 SOQ58016.1 PZC77104.1 PCG77147.1 PZC77116.1 NEVH01025074 PNF16189.1 KQ976745 KYM75416.1 GL887844 EGI69874.1 QOIP01000011 RLU16997.1 KK107021 EZA62323.1 ADTU01022687 GECZ01016734 JAS53035.1 GEBQ01003218 GEBQ01000810 JAT36759.1 JAT39167.1 KK107139 QOIP01000003 EZA57781.1 RLU25049.1 AGBW02010035 OWR49315.1 GBRD01014700 GBRD01014699 JAG51127.1 GDHC01012231 JAQ06398.1 GBRD01015344 JAG50482.1 GBYB01005985 JAG75752.1 GBHO01006478 JAG37126.1 KK853179 KDR10227.1 GEZM01078913 JAV62581.1 GALA01001167 JAA93685.1 AGBW02012985 OWR44136.1 KK854219 PTY13264.1 GEBQ01010386 JAT29591.1 GECZ01014365 JAS55404.1 GEBQ01002115 JAT37862.1 GFDL01005848 JAV29197.1 AB549994 BAI83415.1 KA646086 AFP60715.1 JRES01000423 KNC31408.1 GEBQ01015167 JAT24810.1 JXUM01071369 JXUM01071370 JXUM01071371 JXUM01091985 KQ563956 KQ562660 KXJ73022.1 KXJ75379.1 ODYU01012350 SOQ58622.1 AB901068 BAQ02368.1 GGFK01000934 MBW34255.1 AB901052 BAQ02352.1 GGFK01000993 MBW34314.1 AXCM01000233 AXCN02000090 ADMH02002130 ETN58528.1 KK852809 KDR15985.1 GFDL01005255 JAV29790.1 GFDL01005309 JAV29736.1 GFDL01005268 JAV29777.1 GFDL01005283 JAV29762.1 GEDC01001575 JAS35723.1 EZA57914.1 GECZ01014010 JAS55759.1 GAMD01002846 JAA98744.1 KQ459078 KPJ03807.1 GECU01018986 JAS88720.1 CH477665 EAT37544.1 AAAB01008820 EAA45316.3 EAA45318.1 EAA45319.1 EGK96331.1 KK852657 KDR19246.1 JXJN01006917 GDKW01002359 JAI54236.1 PYGN01001954 PSN32011.1
PCG77150.1 ODYU01011381 SOQ57088.1 LADI01012895 KPJ20745.1 LADJ01037929 KPJ21328.1 NWSH01000558 PCG75665.1 RSAL01000025 RVE52106.1 BABH01040660 KQ459460 KPJ00770.1 PZC77103.1 KPJ00771.1 PZC77105.1 ODYU01001454 SOQ37676.1 PCG75662.1 KPJ00768.1 KPJ00767.1 RVE52115.1 ODYU01012723 SOQ59169.1 PCG75663.1 KQ459838 KPJ19748.1 AGBW02009924 OWR49569.1 KPJ20744.1 PCG75666.1 PZC77102.1 RVE52104.1 BABH01043549 ODYU01011969 SOQ58016.1 PZC77104.1 PCG77147.1 PZC77116.1 NEVH01025074 PNF16189.1 KQ976745 KYM75416.1 GL887844 EGI69874.1 QOIP01000011 RLU16997.1 KK107021 EZA62323.1 ADTU01022687 GECZ01016734 JAS53035.1 GEBQ01003218 GEBQ01000810 JAT36759.1 JAT39167.1 KK107139 QOIP01000003 EZA57781.1 RLU25049.1 AGBW02010035 OWR49315.1 GBRD01014700 GBRD01014699 JAG51127.1 GDHC01012231 JAQ06398.1 GBRD01015344 JAG50482.1 GBYB01005985 JAG75752.1 GBHO01006478 JAG37126.1 KK853179 KDR10227.1 GEZM01078913 JAV62581.1 GALA01001167 JAA93685.1 AGBW02012985 OWR44136.1 KK854219 PTY13264.1 GEBQ01010386 JAT29591.1 GECZ01014365 JAS55404.1 GEBQ01002115 JAT37862.1 GFDL01005848 JAV29197.1 AB549994 BAI83415.1 KA646086 AFP60715.1 JRES01000423 KNC31408.1 GEBQ01015167 JAT24810.1 JXUM01071369 JXUM01071370 JXUM01071371 JXUM01091985 KQ563956 KQ562660 KXJ73022.1 KXJ75379.1 ODYU01012350 SOQ58622.1 AB901068 BAQ02368.1 GGFK01000934 MBW34255.1 AB901052 BAQ02352.1 GGFK01000993 MBW34314.1 AXCM01000233 AXCN02000090 ADMH02002130 ETN58528.1 KK852809 KDR15985.1 GFDL01005255 JAV29790.1 GFDL01005309 JAV29736.1 GFDL01005268 JAV29777.1 GFDL01005283 JAV29762.1 GEDC01001575 JAS35723.1 EZA57914.1 GECZ01014010 JAS55759.1 GAMD01002846 JAA98744.1 KQ459078 KPJ03807.1 GECU01018986 JAS88720.1 CH477665 EAT37544.1 AAAB01008820 EAA45316.3 EAA45318.1 EAA45319.1 EGK96331.1 KK852657 KDR19246.1 JXJN01006917 GDKW01002359 JAI54236.1 PYGN01001954 PSN32011.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000283053
UP000007151
+ More
UP000235965 UP000078540 UP000007755 UP000279307 UP000053097 UP000005205 UP000027135 UP000095301 UP000037069 UP000069940 UP000249989 UP000192223 UP000075883 UP000075886 UP000000673 UP000075902 UP000005203 UP000075900 UP000008820 UP000075882 UP000007062 UP000095300 UP000075903 UP000075920 UP000092460 UP000076407 UP000245037
UP000235965 UP000078540 UP000007755 UP000279307 UP000053097 UP000005205 UP000027135 UP000095301 UP000037069 UP000069940 UP000249989 UP000192223 UP000075883 UP000075886 UP000000673 UP000075902 UP000005203 UP000075900 UP000008820 UP000075882 UP000007062 UP000095300 UP000075903 UP000075920 UP000092460 UP000076407 UP000245037
Pfam
Interpro
IPR005828
MFS_sugar_transport-like
+ More
IPR036259 MFS_trans_sf
IPR005829 Sugar_transporter_CS
IPR020846 MFS_dom
IPR011701 MFS
IPR003663 Sugar/inositol_transpt
IPR007246 Gaa1
IPR036064 MYSc_Myo18
IPR027401 Myosin_IQ_contain_sf
IPR000048 IQ_motif_EF-hand-BS
IPR002928 Myosin_tail
IPR027417 P-loop_NTPase
IPR001609 Myosin_head_motor_dom
IPR036961 Kinesin_motor_dom_sf
IPR036259 MFS_trans_sf
IPR005829 Sugar_transporter_CS
IPR020846 MFS_dom
IPR011701 MFS
IPR003663 Sugar/inositol_transpt
IPR007246 Gaa1
IPR036064 MYSc_Myo18
IPR027401 Myosin_IQ_contain_sf
IPR000048 IQ_motif_EF-hand-BS
IPR002928 Myosin_tail
IPR027417 P-loop_NTPase
IPR001609 Myosin_head_motor_dom
IPR036961 Kinesin_motor_dom_sf
Gene 3D
ProteinModelPortal
H9JVT9
A0A2W1BV94
A0A2A4K0E5
A0A2H1WVK7
A0A0N1IQK5
A0A0N1PKK0
+ More
A0A2A4JUG4 A0A3S2LR68 H9JVU1 A0A194Q6Z4 A0A2W1BZ84 A0A194QBG3 A0A2W1BTX0 A0A2H1VA59 A0A2A4JW98 A0A194Q5E7 A0A194Q5E1 A0A3S2LEN2 A0A2H1X1H2 A0A2A4JW00 A0A0N0PEG4 A0A212F787 A0A0N0PF14 A0A2A4JV69 A0A2W1BPY0 A0A3S2M6I8 H9JWY9 A0A2H1WYH9 A0A2W1BTZ9 A0A2A4JYQ5 A0A2W1BWR1 A0A2J7PIQ0 A0A195AT95 F4W7K1 A0A3L8D924 A0A026X2K8 A0A158NPP3 A0A1B6FS48 A0A1B6MT94 A0A026WQ15 A0A212F6E9 A0A0K8SCV7 A0A146LDU6 A0A0K8SB18 A0A0C9RCE9 A0A0A9YZV0 A0A067QP79 A0A1Y1KQ78 T1DE67 A0A212ERK6 A0A2R7W0J8 A0A1B6M0Z3 A0A1B6FYZ7 A0A1B6MPN8 A0A1Q3FNQ8 A0A1I8NJI3 D4AHW6 T1PBE5 A0A0L0CG94 A0A1B6LM42 A0A182GVZ2 A0A2H1X046 A0A0A8J8K5 A0A2M4A0I2 A0A1W4XL19 A0A0A8J7X6 A0A2M4A0H9 A0A182MM04 A0A182QT48 W5J543 A0A067QZV9 A0A182TSI6 A0A1Q3FQA6 A0A1Q3FQG8 A0A088ABY6 A0A1Q3FQD3 A0A1Q3FQH2 A0A1B6ECS3 A0A182S579 A0A026WS40 A0A1B6FZZ0 T1DG93 A0A1W4XAC6 A0A0N1PEX2 A0A1B6IP97 Q16SU4 A0A182KX77 A0NBZ0 A0A1I8P6M8 A0A067R7D8 A0A182VIR9 A0A182VS58 A0A1B0B100 A0A1Y9J057 A0A0P4VMZ3 A0A2P8XJ31
A0A2A4JUG4 A0A3S2LR68 H9JVU1 A0A194Q6Z4 A0A2W1BZ84 A0A194QBG3 A0A2W1BTX0 A0A2H1VA59 A0A2A4JW98 A0A194Q5E7 A0A194Q5E1 A0A3S2LEN2 A0A2H1X1H2 A0A2A4JW00 A0A0N0PEG4 A0A212F787 A0A0N0PF14 A0A2A4JV69 A0A2W1BPY0 A0A3S2M6I8 H9JWY9 A0A2H1WYH9 A0A2W1BTZ9 A0A2A4JYQ5 A0A2W1BWR1 A0A2J7PIQ0 A0A195AT95 F4W7K1 A0A3L8D924 A0A026X2K8 A0A158NPP3 A0A1B6FS48 A0A1B6MT94 A0A026WQ15 A0A212F6E9 A0A0K8SCV7 A0A146LDU6 A0A0K8SB18 A0A0C9RCE9 A0A0A9YZV0 A0A067QP79 A0A1Y1KQ78 T1DE67 A0A212ERK6 A0A2R7W0J8 A0A1B6M0Z3 A0A1B6FYZ7 A0A1B6MPN8 A0A1Q3FNQ8 A0A1I8NJI3 D4AHW6 T1PBE5 A0A0L0CG94 A0A1B6LM42 A0A182GVZ2 A0A2H1X046 A0A0A8J8K5 A0A2M4A0I2 A0A1W4XL19 A0A0A8J7X6 A0A2M4A0H9 A0A182MM04 A0A182QT48 W5J543 A0A067QZV9 A0A182TSI6 A0A1Q3FQA6 A0A1Q3FQG8 A0A088ABY6 A0A1Q3FQD3 A0A1Q3FQH2 A0A1B6ECS3 A0A182S579 A0A026WS40 A0A1B6FZZ0 T1DG93 A0A1W4XAC6 A0A0N1PEX2 A0A1B6IP97 Q16SU4 A0A182KX77 A0NBZ0 A0A1I8P6M8 A0A067R7D8 A0A182VIR9 A0A182VS58 A0A1B0B100 A0A1Y9J057 A0A0P4VMZ3 A0A2P8XJ31
PDB
5EQI
E-value=8.45643e-17,
Score=214
Ontologies
GO
PANTHER
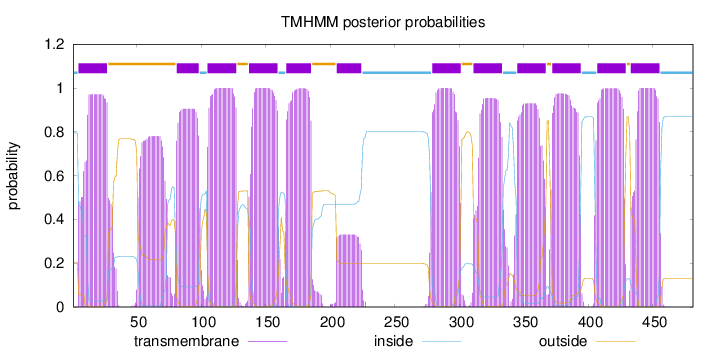
Topology
Length:
481
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
257.60165
Exp number, first 60 AAs:
29.1736
Total prob of N-in:
0.79515
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 80
TMhelix
81 - 98
inside
99 - 104
TMhelix
105 - 127
outside
128 - 136
TMhelix
137 - 159
inside
160 - 165
TMhelix
166 - 185
outside
186 - 204
TMhelix
205 - 224
inside
225 - 278
TMhelix
279 - 301
outside
302 - 310
TMhelix
311 - 333
inside
334 - 344
TMhelix
345 - 367
outside
368 - 371
TMhelix
372 - 394
inside
395 - 406
TMhelix
407 - 429
outside
430 - 432
TMhelix
433 - 455
inside
456 - 481
Population Genetic Test Statistics
Pi
264.128428
Theta
217.007806
Tajima's D
0.839981
CLR
0.008099
CSRT
0.615519224038798
Interpretation
Uncertain
