Gene
KWMTBOMO11791
Pre Gene Modal
BGIBMGA013653
Annotation
PREDICTED:_glycosylphosphatidylinositol_anchor_attachment_1_protein_isoform_X1_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 2.8
Sequence
CDS
ATGGGGCTTTTGTCAGACCCAGAGTACGGTTCGGTAAAATGGGTGAGGATTCTAAAGAAGGTTCACAGGCCGCTTTGCTTCCTTTGTTATGCTGTGGCCCTAGTATGGTTCGTGATGTTGGCTAACAGAGACTTTAATAATGAGACTTATTTTTCTGAAAATGCTCTGCTTCCTGGGCTCGTTACCAACGAGTTCAACGGCGAGTATGCTGCAAAGCAGTATTTGACTGAATTCGAGCAGGAACTGGAGGAAAAGTATTATGACACAGAGAAGATACCAGTCCCGTGGCTGGTGGCCAAGATGTCTCAGCTTCACCTAGAGGTTTACACACACAACTATACACTAAACTACCCTTTAGGACAAGGTCAGATTTACAAAGGCACAAATGTGTATGGCATCCTTAGAGCTGCTCGTACTCCTTCCTTGGAAGCTCTAGTAGTTTCTGCTCCATTCAGACCCCTGTCTTCACATCAGAAGAGCACAGCAGCTGGAATTGCATTAATGTTGGCTTTTGCAGAGTTTGCTAGACCACAAAAATATTGGGCCAAAGATATAATCTTCCTTGTAACGGAACATGAGCAACTAGGAATGCAGGCATGGCTAGAAGCTTATCATGGCCTGCAAAGTGACTCTGAAACATTTTACACTACTCTCGGCAGTTTTTGGAAGCCTGGTAGTTTTGGTCTTGACCTGACTAATTTTTTCCCTGGATTTGACTGGCTTGGGAGTCACAAGAAGTGTCTCAACCCTGGACAGTTGAAAGCCAGGTCTGGAGCAATTCAAGCAGCTATTAATTTGGAGTTCCATTCTCCAAAGATAAAATATGTTGAAGTCAAAGTGGAAGGTTTGAATGGACAACTACCAAACTTGGACCTGGTCAACTTGGTGCACAAGCTGTGTGTGAAGTCCGGTATACACCACTCTTACAAGAACAGCTACTCCTGGGTCCGCAACCGAAATAAAGTGGACGACTGGGTGAACAACATCTGGACCATGGTGGGGATGGTCAGCACGCAACTGGCCGGGGTCCCCAACGGTAACCACGGGCTGTTCCACCGGTTCGGCATAGAGGCCGTGACCCTGGAGGGCCGGGACTCCTCGGACGGGCCGGCGCCGCGCGTGCACTCGCTCTCCGCCGCTAACTTCCACCGCCTCGGCCTCGCGCTCGAGACCGTCCTCCGCTCCCTCAACAACTTGCTGGAGGCCTTCCACCAGAGCTACTTCTTCTACATGCTGCCCGCCACCGATCGGTTTGTGTCCATCGGCCAGTACATGCCCAGCCTGTGCCTGATGTGCGCGGCCATGCTGCTCCGCGCGCTGGCGCTGTGGGTGGGCGTCCAGCCGACACAGGAACGCAGCAGTCAAGCTGAAGACCCCGTAGACTCGGAGGCCAAAGAACCAGGAGAGCGACCGCCTGAACGACTATCAGAAAGCTCAAACCAGCGAGTGCACAACAGGAAGACGATCAAAGAAAAACATAGTGATTCTGAATCTGAGATACCAGGAGAACAGGATGTTAAAGAAGTAGTACAGAGCTCAGACGAAAGGGTCGCGAACAGCCAAACCCAACTAAACTTCAGCATGGTCAATGTCGGTGCCAACTATCTGTTGGTGCACGTGCTGGGGTACGCCGCCATGAACTCGCCCGTCCTCTTCACGCAGCTCGGCGCCAAATACTTTAGTCTGCCCTCCGAGGTGTCGGTGTACTACGGCCTGTGCGTGACCTCCGCCGTGCTGGTGGTGGCCGCGCCCGCCCTGCCGCGCCTGCTCCGCCGCGGCGCCCTCACGCACGAGGAGCTGACGCTGGTCAACATCCTGGTGCTGATCGAGCTGTCGACGCTGTGCCTCGCGCTGGGCATGCACAACTTCCCGCTGGGGCTGGCGGTCGCGGCCCTCTACACGCCGGTCGCCCTGCTGTCAGGAGTTGTAGTCCGAGAACGGGGCTGGATAAGCGTGTGGCTGCAGCGTGCTGCGTGCGCCGTGCTGCAGCCCCTGGGCGCCCTGACCGCCGCCATGTGCGTGTACAGCGTCGCCGTGTACGGGGGGGCCGAGGGGGCCGCAGCCGCCGCGGGGAGGGGCCGGGACGCCGCCATGCAGGCCGTCATGTTCGCCGTGGCAGATTCCCTGATCTACGGCAACTGGGTGTTCAACGTGGCGGCGTCGGTGCTGCTGCCGGTGTGGGTGGTGCTCTGGCAGATCGTCTGCAACAGCGTGGAGGGGCCTCCCCCCGCGCGCCGTTGA
Protein
MGLLSDPEYGSVKWVRILKKVHRPLCFLCYAVALVWFVMLANRDFNNETYFSENALLPGLVTNEFNGEYAAKQYLTEFEQELEEKYYDTEKIPVPWLVAKMSQLHLEVYTHNYTLNYPLGQGQIYKGTNVYGILRAARTPSLEALVVSAPFRPLSSHQKSTAAGIALMLAFAEFARPQKYWAKDIIFLVTEHEQLGMQAWLEAYHGLQSDSETFYTTLGSFWKPGSFGLDLTNFFPGFDWLGSHKKCLNPGQLKARSGAIQAAINLEFHSPKIKYVEVKVEGLNGQLPNLDLVNLVHKLCVKSGIHHSYKNSYSWVRNRNKVDDWVNNIWTMVGMVSTQLAGVPNGNHGLFHRFGIEAVTLEGRDSSDGPAPRVHSLSAANFHRLGLALETVLRSLNNLLEAFHQSYFFYMLPATDRFVSIGQYMPSLCLMCAAMLLRALALWVGVQPTQERSSQAEDPVDSEAKEPGERPPERLSESSNQRVHNRKTIKEKHSDSESEIPGEQDVKEVVQSSDERVANSQTQLNFSMVNVGANYLLVHVLGYAAMNSPVLFTQLGAKYFSLPSEVSVYYGLCVTSAVLVVAAPALPRLLRRGALTHEELTLVNILVLIELSTLCLALGMHNFPLGLAVAALYTPVALLSGVVVRERGWISVWLQRAACAVLQPLGALTAAMCVYSVAVYGGAEGAAAAAGRGRDAAMQAVMFAVADSLIYGNWVFNVAASVLLPVWVVLWQIVCNSVEGPPPARR
Summary
Uniprot
H9JVU0
A0A2W1BWR1
A0A2A4JZN8
A0A194Q6Y9
A0A1E1WKS3
A0A3S2NNF2
+ More
A0A2H1VTF8 D6WQR2 A0A026W1Q4 A0A067RR79 A0A158NVM6 E2BWN5 A0A195B7P1 A0A195ED68 A0A195FV43 A0A151XBX5 F4WNQ6 A0A1Y1M877 A0A151IP35 A0A232FGX8 A0A0M8ZQR3 A0A2J7RLZ5 A0A2P8ZCF1 A0A0L7QNZ5 A0A1B6C667 K7IR57 A0A154P008 A0A182KV50 A0A182I5W5 V9IEU6 A0A182WXU4 Q7QDZ1 U5EPK2 A0A182VDG6 A0A336MQS1 A0A1A9WAP1
A0A2H1VTF8 D6WQR2 A0A026W1Q4 A0A067RR79 A0A158NVM6 E2BWN5 A0A195B7P1 A0A195ED68 A0A195FV43 A0A151XBX5 F4WNQ6 A0A1Y1M877 A0A151IP35 A0A232FGX8 A0A0M8ZQR3 A0A2J7RLZ5 A0A2P8ZCF1 A0A0L7QNZ5 A0A1B6C667 K7IR57 A0A154P008 A0A182KV50 A0A182I5W5 V9IEU6 A0A182WXU4 Q7QDZ1 U5EPK2 A0A182VDG6 A0A336MQS1 A0A1A9WAP1
Pubmed
EMBL
BABH01040661
KZ149937
PZC77116.1
NWSH01000350
PCG77148.1
KQ459460
+ More
KPJ00765.1 GDQN01003526 JAT87528.1 RSAL01000025 RVE52116.1 ODYU01004180 SOQ43772.1 KQ971354 EFA06039.1 KK107558 QOIP01000012 EZA48989.1 RLU15978.1 KK852513 KDR22254.1 ADTU01002895 GL451165 EFN79890.1 KQ976565 KYM80543.1 KQ979074 KYN22789.1 KQ981215 KYN44510.1 KQ982316 KYQ57847.1 GL888237 EGI64249.1 GEZM01039758 JAV81218.1 KQ976892 KYN07229.1 NNAY01000242 OXU29770.1 KQ435955 KOX68046.1 NEVH01002601 PNF41829.1 PYGN01000106 PSN54175.1 KQ414843 KOC60304.1 GEDC01028578 GEDC01008103 JAS08720.1 JAS29195.1 KQ434786 KZC05172.1 APCN01003535 JR042544 AEY59633.1 AAAB01008848 EAA07058.4 GANO01000138 JAB59733.1 UFQT01002145 SSX32762.1
KPJ00765.1 GDQN01003526 JAT87528.1 RSAL01000025 RVE52116.1 ODYU01004180 SOQ43772.1 KQ971354 EFA06039.1 KK107558 QOIP01000012 EZA48989.1 RLU15978.1 KK852513 KDR22254.1 ADTU01002895 GL451165 EFN79890.1 KQ976565 KYM80543.1 KQ979074 KYN22789.1 KQ981215 KYN44510.1 KQ982316 KYQ57847.1 GL888237 EGI64249.1 GEZM01039758 JAV81218.1 KQ976892 KYN07229.1 NNAY01000242 OXU29770.1 KQ435955 KOX68046.1 NEVH01002601 PNF41829.1 PYGN01000106 PSN54175.1 KQ414843 KOC60304.1 GEDC01028578 GEDC01008103 JAS08720.1 JAS29195.1 KQ434786 KZC05172.1 APCN01003535 JR042544 AEY59633.1 AAAB01008848 EAA07058.4 GANO01000138 JAB59733.1 UFQT01002145 SSX32762.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000007266
UP000053097
+ More
UP000279307 UP000027135 UP000005205 UP000008237 UP000078540 UP000078492 UP000078541 UP000075809 UP000007755 UP000078542 UP000215335 UP000053105 UP000235965 UP000245037 UP000053825 UP000002358 UP000076502 UP000075882 UP000075840 UP000076407 UP000007062 UP000075903 UP000091820
UP000279307 UP000027135 UP000005205 UP000008237 UP000078540 UP000078492 UP000078541 UP000075809 UP000007755 UP000078542 UP000215335 UP000053105 UP000235965 UP000245037 UP000053825 UP000002358 UP000076502 UP000075882 UP000075840 UP000076407 UP000007062 UP000075903 UP000091820
PRIDE
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JVU0
A0A2W1BWR1
A0A2A4JZN8
A0A194Q6Y9
A0A1E1WKS3
A0A3S2NNF2
+ More
A0A2H1VTF8 D6WQR2 A0A026W1Q4 A0A067RR79 A0A158NVM6 E2BWN5 A0A195B7P1 A0A195ED68 A0A195FV43 A0A151XBX5 F4WNQ6 A0A1Y1M877 A0A151IP35 A0A232FGX8 A0A0M8ZQR3 A0A2J7RLZ5 A0A2P8ZCF1 A0A0L7QNZ5 A0A1B6C667 K7IR57 A0A154P008 A0A182KV50 A0A182I5W5 V9IEU6 A0A182WXU4 Q7QDZ1 U5EPK2 A0A182VDG6 A0A336MQS1 A0A1A9WAP1
A0A2H1VTF8 D6WQR2 A0A026W1Q4 A0A067RR79 A0A158NVM6 E2BWN5 A0A195B7P1 A0A195ED68 A0A195FV43 A0A151XBX5 F4WNQ6 A0A1Y1M877 A0A151IP35 A0A232FGX8 A0A0M8ZQR3 A0A2J7RLZ5 A0A2P8ZCF1 A0A0L7QNZ5 A0A1B6C667 K7IR57 A0A154P008 A0A182KV50 A0A182I5W5 V9IEU6 A0A182WXU4 Q7QDZ1 U5EPK2 A0A182VDG6 A0A336MQS1 A0A1A9WAP1
Ontologies
PATHWAY
GO
PANTHER
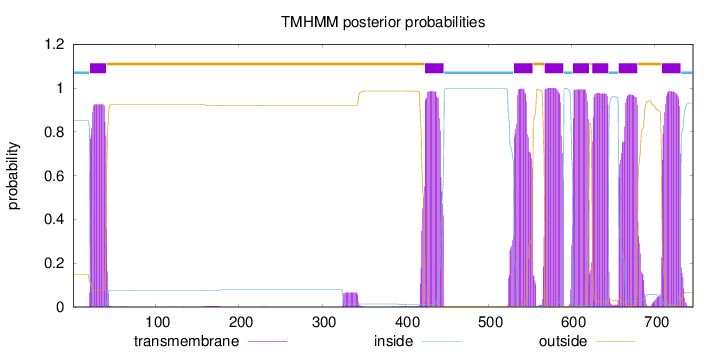
Topology
Length:
746
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
171.44394
Exp number, first 60 AAs:
18.48848
Total prob of N-in:
0.85095
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 40
outside
41 - 423
TMhelix
424 - 446
inside
447 - 530
TMhelix
531 - 553
outside
554 - 567
TMhelix
568 - 590
inside
591 - 601
TMhelix
602 - 621
outside
622 - 624
TMhelix
625 - 644
inside
645 - 656
TMhelix
657 - 679
outside
680 - 708
TMhelix
709 - 731
inside
732 - 746
Population Genetic Test Statistics
Pi
249.897042
Theta
206.641053
Tajima's D
0.749772
CLR
0.003765
CSRT
0.592120393980301
Interpretation
Uncertain
