Gene
KWMTBOMO11790
Pre Gene Modal
BGIBMGA013654
Annotation
PREDICTED:_monosaccharide-sensing_protein_3-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.901
Sequence
CDS
ATGGCTGCGATGTTCATGTGGCCATCCTCTACGCTGACACTCTTCCAATGCGACAACACACCGCTGCATCGCCCCTTGACGGAGGCCGAGGCGAACATGCTAGGCAGCCTGTCCTCTCTGGGCGCTCTCATCAGCACGCCCTTCACTGGAATGATGCTCGACCGACTCGGTAGGAAGAACATATCCTTGCTCATAGGAGTTATAGGTGTGATATCGTGGTCAATGATTTCCTTGTCCAACCGGGTAGAAGTGGTTCTGGTCGCCATTTTCTTGAGCGGCCTCAGCGGCTCCGTGTTCCTAGTAGTTCCAGTCTACATCTCTGAGATATGTCAGACATCGATTCGAGGGACGATGACCTCTTGCTCCATGGTGTTCTACGGCGTGGGCATGTTGGTCTCGTACCTGATGGGCGGCTACATGAACTACTACCCCATGGTGTACACGTGTCTGACCATGGCAGTAGTTAGTACACTCTTCATTACCGTCCTAAAGGAAACTCCGGTTTGTCTGATGGCTAAGGGATTAAACGAGGAAGCAGCCAAGTCGTTGGCGTTTTATAGAGGACTAAAACCAGAGTCTAAAGAAGTGTTGGAAGAGTTGAACATCATTCAGAGATCACTGTCTCCTGACTTGGACTGTGCAGAAGTGACTCCTGAGGAACAAAAACTGAATCCGGCCGACAGAAAACATCAAAAATTATCTGTGATACAGTTCATGAACAAATCAAGATCTACGCGCAAAGCTCTGTTCGTCTGCGTCACGATATTGACACTGTCAATATTCCAAGGATCGGTGGTGGTACAAGTGTACGCGGGGCCCTTATTCTCTGCGGCGATCCCGTCCGTGTCGTCCACTCTGTGCAGCGTAGTGGTCGCCGTGGTCACAGTCCTCACCGGGCTGGTCTCCGCCTACTTGACTGAGGTCACCGGAAGACGACCGCTGATGATCCACTCGTCCACACTGAGCGGGTTGTGCTGTATCCTCCTCGGTACTCAAATACAGCTGGGCTGGGCTCCGAGCTGGGTGACTGCCCTCTTCATCTTTGTCTTCACCGCGACGTACAGCCTCGGCTCCGGCATGGTGCCGTACGTGCTGTTTGCTGAAGTATTCTTGCCCGAGGTGAAGAGCTTCATGTCGATGCTGACGGTGGAATGGGCCTGGTTCTGCAACTACGTGATCCTGTTGATCTTCAACCCGCTGCTCAAGGTCGTCGGTCTGGGTCCGATTTTTTACATATTCGCGGTCATCTGCTTCGTGACCAGCGTGTTCTGTATGCTGGAGCAGCCGGAGACCAAGGGCCTCCATTGCGAAGACATTCAACACTTATTCGATAAGAAGAGCAGTAAGGCGCGTCGGATCGAGGGCTCTGCGTGA
Protein
MAAMFMWPSSTLTLFQCDNTPLHRPLTEAEANMLGSLSSLGALISTPFTGMMLDRLGRKNISLLIGVIGVISWSMISLSNRVEVVLVAIFLSGLSGSVFLVVPVYISEICQTSIRGTMTSCSMVFYGVGMLVSYLMGGYMNYYPMVYTCLTMAVVSTLFITVLKETPVCLMAKGLNEEAAKSLAFYRGLKPESKEVLEELNIIQRSLSPDLDCAEVTPEEQKLNPADRKHQKLSVIQFMNKSRSTRKALFVCVTILTLSIFQGSVVVQVYAGPLFSAAIPSVSSTLCSVVVAVVTVLTGLVSAYLTEVTGRRPLMIHSSTLSGLCCILLGTQIQLGWAPSWVTALFIFVFTATYSLGSGMVPYVLFAEVFLPEVKSFMSMLTVEWAWFCNYVILLIFNPLLKVVGLGPIFYIFAVICFVTSVFCMLEQPETKGLHCEDIQHLFDKKSSKARRIEGSA
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9JVU1
A0A2W1BTX0
A0A2A4JW98
A0A194Q5E7
A0A2H1VA59
A0A194Q5E1
+ More
A0A3S2LEN2 A0A194QBG3 A0A194Q6Z4 A0A2A4JYQ5 A0A0N0PEG4 A0A2W1BWR1 A0A0L7L2W9 A0A0N1PKK0 A0A0N1IQK5 A0A2A4JUG4 A0A2W1BZ84 A0A212F787 A0A2A4JV69 A0A2W1BPY0 A0A3S2M6I8 A0A2H1X1H2 A0A2H1WYH9 H9JWY9 A0A3S2LR68 H9JWZ0 A0A212F6E9 A0A2A4JW00 A0A0N0PF14 A0A2W1BTZ9 A0A2A4K0E5 A0A2W1BV94 A0A2J7R8M9 A0A2H1WVK7 A0A0N1PIZ3 A0A2J7PIQ0 A0A212F505 A0A2J7PYX6 A0A2P8YT21 A0A1I8NPR8 A0A2H1X046 A0A1I8NPR6 D4AHW9 A0A3B0JHR8 A0A2A4J9N1 A0A1B6GYS0 A0A2P8ZDK3 A0A084W7N7 A0A1Y1KQ78 A0A0N8P1N8 B3LWC6 A0A1L8DFE1 A0A1L8DF45 A0A226F053 A0A336LI37 A0A226DUM5 E9IQR5 U5EQL0 A0A2W1BU14 I5AP16 A0A1Q3FJP9 B5DW12 B4G347 A0A1L8DFB2 A0A1Q3FJI4 A0A1B0CBU8 D6X0L8 A0A2A4JKE4 A0A1Y9H2S4 W8ATJ1 Q16ZD3 B0X8T3 A0A1S4F5X0 B4LXB4 B4JH51 Q17EH6 A0A0Q9WS19 A0A1B0CBU7 A0A2H1WM77 A0A182QQ42 A0A1B6FS48 A0A0N1IGQ6 A0A182T8Z5 A0A1A9VZI8 A0A2A4JPP7 A0A2A4K8F1 A0A182P8W6 A0A0M3QYJ6 A0A2A4K8N6 A0A1B0G9W5 A0A1L8DIV6
A0A3S2LEN2 A0A194QBG3 A0A194Q6Z4 A0A2A4JYQ5 A0A0N0PEG4 A0A2W1BWR1 A0A0L7L2W9 A0A0N1PKK0 A0A0N1IQK5 A0A2A4JUG4 A0A2W1BZ84 A0A212F787 A0A2A4JV69 A0A2W1BPY0 A0A3S2M6I8 A0A2H1X1H2 A0A2H1WYH9 H9JWY9 A0A3S2LR68 H9JWZ0 A0A212F6E9 A0A2A4JW00 A0A0N0PF14 A0A2W1BTZ9 A0A2A4K0E5 A0A2W1BV94 A0A2J7R8M9 A0A2H1WVK7 A0A0N1PIZ3 A0A2J7PIQ0 A0A212F505 A0A2J7PYX6 A0A2P8YT21 A0A1I8NPR8 A0A2H1X046 A0A1I8NPR6 D4AHW9 A0A3B0JHR8 A0A2A4J9N1 A0A1B6GYS0 A0A2P8ZDK3 A0A084W7N7 A0A1Y1KQ78 A0A0N8P1N8 B3LWC6 A0A1L8DFE1 A0A1L8DF45 A0A226F053 A0A336LI37 A0A226DUM5 E9IQR5 U5EQL0 A0A2W1BU14 I5AP16 A0A1Q3FJP9 B5DW12 B4G347 A0A1L8DFB2 A0A1Q3FJI4 A0A1B0CBU8 D6X0L8 A0A2A4JKE4 A0A1Y9H2S4 W8ATJ1 Q16ZD3 B0X8T3 A0A1S4F5X0 B4LXB4 B4JH51 Q17EH6 A0A0Q9WS19 A0A1B0CBU7 A0A2H1WM77 A0A182QQ42 A0A1B6FS48 A0A0N1IGQ6 A0A182T8Z5 A0A1A9VZI8 A0A2A4JPP7 A0A2A4K8F1 A0A182P8W6 A0A0M3QYJ6 A0A2A4K8N6 A0A1B0G9W5 A0A1L8DIV6
Pubmed
EMBL
BABH01040660
KZ149937
PZC77105.1
NWSH01000558
PCG75662.1
KQ459460
+ More
KPJ00768.1 ODYU01001454 SOQ37676.1 KPJ00767.1 RSAL01000025 RVE52115.1 KPJ00771.1 KPJ00770.1 NWSH01000350 PCG77147.1 KQ459838 KPJ19748.1 PZC77116.1 JTDY01003373 KOB69656.1 LADJ01037929 KPJ21328.1 LADI01012895 KPJ20745.1 PCG75665.1 PZC77103.1 AGBW02009924 OWR49569.1 PCG75666.1 PZC77102.1 RVE52104.1 ODYU01012723 SOQ59169.1 ODYU01011969 SOQ58016.1 BABH01043549 RVE52106.1 BABH01041799 BABH01041800 BABH01041801 AGBW02010035 OWR49315.1 PCG75663.1 KPJ20744.1 PZC77104.1 PCG77150.1 PZC77117.1 NEVH01006721 PNF37175.1 ODYU01011381 SOQ57088.1 KQ460009 KPJ18482.1 NEVH01025074 PNF16189.1 AGBW02010281 OWR48812.1 NEVH01020342 PNF21526.1 PYGN01000377 PSN47391.1 ODYU01012350 SOQ58622.1 AB549997 BAI83418.1 OUUW01000005 SPP80253.1 NWSH01002244 PCG68837.1 GECZ01003576 GECZ01002212 JAS66193.1 JAS67557.1 PYGN01000088 PSN54587.1 ATLV01021268 KE525315 KFB46231.1 GEZM01078913 JAV62581.1 CH902617 KPU80444.1 EDV43759.1 GFDF01009004 JAV05080.1 GFDF01009005 JAV05079.1 LNIX01000001 OXA62848.1 UFQT01000014 SSX17682.1 LNIX01000011 OXA48919.1 GL764900 EFZ17072.1 GANO01004191 JAB55680.1 KZ149936 PZC77145.1 CM000070 EIM52701.1 GFDL01007246 JAV27799.1 EDY68099.1 CH479179 EDW24242.1 GFDF01009009 JAV05075.1 GFDL01007265 JAV27780.1 AJWK01005837 AJWK01005838 KQ971371 EFA10112.2 NWSH01001270 PCG71903.1 GAMC01014390 JAB92165.1 CH477492 EAT40014.1 DS232501 EDS42675.1 CH940650 EDW67792.1 CH916369 EDV92742.1 CH477282 EAT44903.1 KRF83470.1 AJWK01005834 AJWK01005835 AJWK01005836 ODYU01009532 SOQ54032.1 AXCN02000705 GECZ01016734 JAS53035.1 KQ459910 KPJ19331.1 NWSH01000916 PCG73554.1 NWSH01000052 PCG80174.1 CP012526 ALC47793.1 PCG80173.1 CCAG010008567 GFDF01007794 JAV06290.1
KPJ00768.1 ODYU01001454 SOQ37676.1 KPJ00767.1 RSAL01000025 RVE52115.1 KPJ00771.1 KPJ00770.1 NWSH01000350 PCG77147.1 KQ459838 KPJ19748.1 PZC77116.1 JTDY01003373 KOB69656.1 LADJ01037929 KPJ21328.1 LADI01012895 KPJ20745.1 PCG75665.1 PZC77103.1 AGBW02009924 OWR49569.1 PCG75666.1 PZC77102.1 RVE52104.1 ODYU01012723 SOQ59169.1 ODYU01011969 SOQ58016.1 BABH01043549 RVE52106.1 BABH01041799 BABH01041800 BABH01041801 AGBW02010035 OWR49315.1 PCG75663.1 KPJ20744.1 PZC77104.1 PCG77150.1 PZC77117.1 NEVH01006721 PNF37175.1 ODYU01011381 SOQ57088.1 KQ460009 KPJ18482.1 NEVH01025074 PNF16189.1 AGBW02010281 OWR48812.1 NEVH01020342 PNF21526.1 PYGN01000377 PSN47391.1 ODYU01012350 SOQ58622.1 AB549997 BAI83418.1 OUUW01000005 SPP80253.1 NWSH01002244 PCG68837.1 GECZ01003576 GECZ01002212 JAS66193.1 JAS67557.1 PYGN01000088 PSN54587.1 ATLV01021268 KE525315 KFB46231.1 GEZM01078913 JAV62581.1 CH902617 KPU80444.1 EDV43759.1 GFDF01009004 JAV05080.1 GFDF01009005 JAV05079.1 LNIX01000001 OXA62848.1 UFQT01000014 SSX17682.1 LNIX01000011 OXA48919.1 GL764900 EFZ17072.1 GANO01004191 JAB55680.1 KZ149936 PZC77145.1 CM000070 EIM52701.1 GFDL01007246 JAV27799.1 EDY68099.1 CH479179 EDW24242.1 GFDF01009009 JAV05075.1 GFDL01007265 JAV27780.1 AJWK01005837 AJWK01005838 KQ971371 EFA10112.2 NWSH01001270 PCG71903.1 GAMC01014390 JAB92165.1 CH477492 EAT40014.1 DS232501 EDS42675.1 CH940650 EDW67792.1 CH916369 EDV92742.1 CH477282 EAT44903.1 KRF83470.1 AJWK01005834 AJWK01005835 AJWK01005836 ODYU01009532 SOQ54032.1 AXCN02000705 GECZ01016734 JAS53035.1 KQ459910 KPJ19331.1 NWSH01000916 PCG73554.1 NWSH01000052 PCG80174.1 CP012526 ALC47793.1 PCG80173.1 CCAG010008567 GFDF01007794 JAV06290.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000053240
UP000037510
+ More
UP000007151 UP000235965 UP000245037 UP000095300 UP000268350 UP000030765 UP000007801 UP000198287 UP000001819 UP000008744 UP000092461 UP000007266 UP000075884 UP000008820 UP000002320 UP000008792 UP000001070 UP000075886 UP000075901 UP000091820 UP000075885 UP000092553 UP000092444
UP000007151 UP000235965 UP000245037 UP000095300 UP000268350 UP000030765 UP000007801 UP000198287 UP000001819 UP000008744 UP000092461 UP000007266 UP000075884 UP000008820 UP000002320 UP000008792 UP000001070 UP000075886 UP000075901 UP000091820 UP000075885 UP000092553 UP000092444
Interpro
IPR020846
MFS_dom
+ More
IPR005829 Sugar_transporter_CS
IPR036259 MFS_trans_sf
IPR005828 MFS_sugar_transport-like
IPR007246 Gaa1
IPR003663 Sugar/inositol_transpt
IPR011701 MFS
IPR013748 Rep_factorC_C
IPR003959 ATPase_AAA_core
IPR003593 AAA+_ATPase
IPR027417 P-loop_NTPase
IPR008921 DNA_pol3_clamp-load_cplx_C
IPR005829 Sugar_transporter_CS
IPR036259 MFS_trans_sf
IPR005828 MFS_sugar_transport-like
IPR007246 Gaa1
IPR003663 Sugar/inositol_transpt
IPR011701 MFS
IPR013748 Rep_factorC_C
IPR003959 ATPase_AAA_core
IPR003593 AAA+_ATPase
IPR027417 P-loop_NTPase
IPR008921 DNA_pol3_clamp-load_cplx_C
CDD
ProteinModelPortal
H9JVU1
A0A2W1BTX0
A0A2A4JW98
A0A194Q5E7
A0A2H1VA59
A0A194Q5E1
+ More
A0A3S2LEN2 A0A194QBG3 A0A194Q6Z4 A0A2A4JYQ5 A0A0N0PEG4 A0A2W1BWR1 A0A0L7L2W9 A0A0N1PKK0 A0A0N1IQK5 A0A2A4JUG4 A0A2W1BZ84 A0A212F787 A0A2A4JV69 A0A2W1BPY0 A0A3S2M6I8 A0A2H1X1H2 A0A2H1WYH9 H9JWY9 A0A3S2LR68 H9JWZ0 A0A212F6E9 A0A2A4JW00 A0A0N0PF14 A0A2W1BTZ9 A0A2A4K0E5 A0A2W1BV94 A0A2J7R8M9 A0A2H1WVK7 A0A0N1PIZ3 A0A2J7PIQ0 A0A212F505 A0A2J7PYX6 A0A2P8YT21 A0A1I8NPR8 A0A2H1X046 A0A1I8NPR6 D4AHW9 A0A3B0JHR8 A0A2A4J9N1 A0A1B6GYS0 A0A2P8ZDK3 A0A084W7N7 A0A1Y1KQ78 A0A0N8P1N8 B3LWC6 A0A1L8DFE1 A0A1L8DF45 A0A226F053 A0A336LI37 A0A226DUM5 E9IQR5 U5EQL0 A0A2W1BU14 I5AP16 A0A1Q3FJP9 B5DW12 B4G347 A0A1L8DFB2 A0A1Q3FJI4 A0A1B0CBU8 D6X0L8 A0A2A4JKE4 A0A1Y9H2S4 W8ATJ1 Q16ZD3 B0X8T3 A0A1S4F5X0 B4LXB4 B4JH51 Q17EH6 A0A0Q9WS19 A0A1B0CBU7 A0A2H1WM77 A0A182QQ42 A0A1B6FS48 A0A0N1IGQ6 A0A182T8Z5 A0A1A9VZI8 A0A2A4JPP7 A0A2A4K8F1 A0A182P8W6 A0A0M3QYJ6 A0A2A4K8N6 A0A1B0G9W5 A0A1L8DIV6
A0A3S2LEN2 A0A194QBG3 A0A194Q6Z4 A0A2A4JYQ5 A0A0N0PEG4 A0A2W1BWR1 A0A0L7L2W9 A0A0N1PKK0 A0A0N1IQK5 A0A2A4JUG4 A0A2W1BZ84 A0A212F787 A0A2A4JV69 A0A2W1BPY0 A0A3S2M6I8 A0A2H1X1H2 A0A2H1WYH9 H9JWY9 A0A3S2LR68 H9JWZ0 A0A212F6E9 A0A2A4JW00 A0A0N0PF14 A0A2W1BTZ9 A0A2A4K0E5 A0A2W1BV94 A0A2J7R8M9 A0A2H1WVK7 A0A0N1PIZ3 A0A2J7PIQ0 A0A212F505 A0A2J7PYX6 A0A2P8YT21 A0A1I8NPR8 A0A2H1X046 A0A1I8NPR6 D4AHW9 A0A3B0JHR8 A0A2A4J9N1 A0A1B6GYS0 A0A2P8ZDK3 A0A084W7N7 A0A1Y1KQ78 A0A0N8P1N8 B3LWC6 A0A1L8DFE1 A0A1L8DF45 A0A226F053 A0A336LI37 A0A226DUM5 E9IQR5 U5EQL0 A0A2W1BU14 I5AP16 A0A1Q3FJP9 B5DW12 B4G347 A0A1L8DFB2 A0A1Q3FJI4 A0A1B0CBU8 D6X0L8 A0A2A4JKE4 A0A1Y9H2S4 W8ATJ1 Q16ZD3 B0X8T3 A0A1S4F5X0 B4LXB4 B4JH51 Q17EH6 A0A0Q9WS19 A0A1B0CBU7 A0A2H1WM77 A0A182QQ42 A0A1B6FS48 A0A0N1IGQ6 A0A182T8Z5 A0A1A9VZI8 A0A2A4JPP7 A0A2A4K8F1 A0A182P8W6 A0A0M3QYJ6 A0A2A4K8N6 A0A1B0G9W5 A0A1L8DIV6
PDB
4YBQ
E-value=1.53946e-20,
Score=246
Ontologies
GO
PANTHER
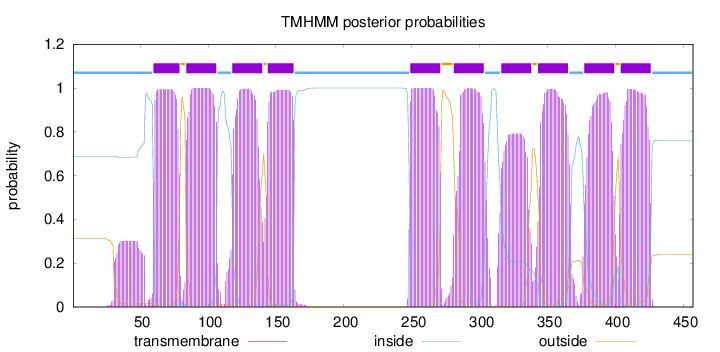
Topology
Length:
457
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
217.68906
Exp number, first 60 AAs:
7.27417
Total prob of N-in:
0.68761
inside
1 - 59
TMhelix
60 - 79
outside
80 - 83
TMhelix
84 - 106
inside
107 - 117
TMhelix
118 - 140
outside
141 - 143
TMhelix
144 - 163
inside
164 - 248
TMhelix
249 - 271
outside
272 - 280
TMhelix
281 - 303
inside
304 - 315
TMhelix
316 - 338
outside
339 - 342
TMhelix
343 - 365
inside
366 - 376
TMhelix
377 - 399
outside
400 - 403
TMhelix
404 - 426
inside
427 - 457
Population Genetic Test Statistics
Pi
314.544281
Theta
199.503945
Tajima's D
1.714059
CLR
0.317402
CSRT
0.833708314584271
Interpretation
Uncertain
