Gene
KWMTBOMO11789 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013655
Annotation
PREDICTED:_agrin_[Papilio_xuthus]
Location in the cell
Extracellular Reliability : 4.066
Sequence
CDS
ATGGAGTCCGGGACCTACCTCCTCGCCGCTGCTGCCCTCCTTGTCCTCGCGCCGACGAGCGAGGCAGCTCGCGACGCGTCGTGTCCTCGCATCTGCGGGCCGGCGCTGCAGGGGGAGCCCGTCTGCGCCACCGACGGGTACATCTACCCCTCGCTCTGCGAGATGAGGAAGAAGACTTGCGGGAAAGGAGTGCGGCTAGCCCCGGACCAGGGCTCATGTTCCCGCGCGCAGGGCTCCAAGTGTGACCACCGCTGCACGTCGGAGCGGGACCCGGTCTGCGGGACCAACGGACGGACCTACCTCAACCGCTGCATGCTGCAGGTCGAAATCTGCAGGCTCGGCATAGGCCTGTCTCACCTGGGCGCCTGCAACAACATCAGCGCGCACCGCGAGAACTGCCCCGTCGACTGCTCGCAGGCCCCGCTCGACGGCCCCATCTGCGGCTCCGACGGCAACGTGTACAAGAGCACGTGCCAGATGAAGCTGCTCACTTGCGGACAAGGAGTGGTCCGCACCAGCAAGAAGCACTGTCAGACGACCCGCCACTGCCGCGAGTCGTGCTGGCGGGCGGCGCGCCCCACCTGCGGCTCGGACGGGAAGCTCTACGCCAACGCGTGTCGCATGAAGGCTACTAATTGCGGCAAGCACGTGTTCGAAGTACCGATGGCGTTCTGCGTGTCACAAGAGCGCACCTCCGGGGGAGAGTCCTGCTCCACCGACTGCTCCGGAGAGAAAGAGAAACCCGTCTGTGGCTCTGATGAAAACATTTACAGGAATGAATGCGAGATGAAGATGCTAAACTGTGGGATAAACAACAGGAAGATGGTGAAGAGGGTGGACATGGAAAAGTGCAAGTCCAAAATGAACAAGTGTCTGAAGGTGAAATGTCCGAGCGACGCGGACCCCGTGTGCGGTACCGACGCTATCGTCTACGCCAACTCCTGCCACTTGAAGGTCGCCACTTGCTTACGAGGAGTTCAGCTGGCTCACTTCGGAAACTGCACGCTTCTGCCGCGCCTGGAAACCGACTGCCCCGACAACTGCGACAACGTGCTGGAGCAGCCCGTCTGCGGATCCGACGGAAACGTCTACAGGTCTGAGTGCGAGCTGCGGCGGTTGACGTGCGGGCAGCACGTGGTGGCGGTGGCGGCGTCGCACTGCCGCACCACGGCGCTCTGCCACGAGCACTGCCCCGACACGCCCGCCTTCATCTGCGGCTCCGACAACCGCTTCTACAAGAACGAGTGCCTCATGAAGAAGGAGAACTGCGGCAAGCACGTGTTCGTGGTGCCGCTGAAGCGCTGCCTGGCGCGGTTCCAGTACGCGGGCTGCGCGCGCGTGTGCCCGCCGGAGTACGACCCGGTCTGCGGGACCGACGACAAGACCTACTCCAACAAGTGCTTCCTCGAAATGGAGAATTGCCGCTCGAGGAGTCTCGTCCAAATGAAGTACCTGGGGACGTGCTCTGAGCCGATCGCGGAGGAGCCCAAGAACTATCTGTATAGAGACGCCGTAGCCTTGCAAGGCTTTGACGGTATATGTATCTGA
Protein
MESGTYLLAAAALLVLAPTSEAARDASCPRICGPALQGEPVCATDGYIYPSLCEMRKKTCGKGVRLAPDQGSCSRAQGSKCDHRCTSERDPVCGTNGRTYLNRCMLQVEICRLGIGLSHLGACNNISAHRENCPVDCSQAPLDGPICGSDGNVYKSTCQMKLLTCGQGVVRTSKKHCQTTRHCRESCWRAARPTCGSDGKLYANACRMKATNCGKHVFEVPMAFCVSQERTSGGESCSTDCSGEKEKPVCGSDENIYRNECEMKMLNCGINNRKMVKRVDMEKCKSKMNKCLKVKCPSDADPVCGTDAIVYANSCHLKVATCLRGVQLAHFGNCTLLPRLETDCPDNCDNVLEQPVCGSDGNVYRSECELRRLTCGQHVVAVAASHCRTTALCHEHCPDTPAFICGSDNRFYKNECLMKKENCGKHVFVVPLKRCLARFQYAGCARVCPPEYDPVCGTDDKTYSNKCFLEMENCRSRSLVQMKYLGTCSEPIAEEPKNYLYRDAVALQGFDGICI
Summary
Uniprot
H9JVU2
A0A3S2PI71
A0A194Q6Q4
A0A2W1BWQ1
A0A2A4J6V9
A0A2H1WHA9
+ More
D6WQS6 A0A1Y1L7A7 A0A0N1IBU2 V9PBG1 A0A0N1IJC7 J9JQ25 A0A2H8TQ10 A0A1B6LAA0 A0A0L7RDZ6 A0A3L8DW56 E2BTI8 A0A151IXG5 V9IBY7 A0A088AN12 A0A158NYA9 A0A2A3EFI1 K7IQL3 A0A0N0U4B0 A0A0C9Q925 A0A1B6CT80 A0A2R7W8D6 A0A026W4W3 F4WDN7 A0A195FWF6 A0A151I649 A0A151WIL4 A0A154PCZ6 A0A067QZ16 A0A2J7RP32 A0A232ENJ4 A0A2K8JMK5 A0A0T6B9T4 A0A310SUB4 A0A1B6JGX8 E0W0F0 A0A0A9YGG1 A0A0P4VNY1 R4G7R1 A0A1V1G1B5 A0A3B0KN88 A0A1D2NDH1 B4KYE2 A0A1A9YIE2 A0A182FHS8 A0A1W4USG1 A0A1A9UG86 B3NFS1 A0A0J9RRA7 Q2LYL4 B4H638 B4HJI5 A0A1A9ZRX2 A0A0M4ELZ0 A0A0K8VC88 B4PG26 A0A0M3QW31 Q9VSK1 B4IZE3 A0A1J1IAV4 A0A182P3D8 B4LDS4 A0A182VJF9 A0A182Y0T7 A0A182K3P1 B3M5I5 A0A336LWW7 A0A182JGB4 A0A084VDL5 A0A182M715 A0A182UCD6 A0A182XCT5 Q7Q780 A0A182HM62 B4N6D8 A0A182QTC8 A0A034V5E2 A0A182NMA6 Q16G53 A0A182RQA6 A0A1B6GXJ8 A0A0C9RQL4 A0A1I8PZR6 A0A182WK74 W8BGU0 B0WYC2 A0A226F6N1 A0A182HEV5 W5JH31 A0A1S4G226 A0A1A9WVW6
D6WQS6 A0A1Y1L7A7 A0A0N1IBU2 V9PBG1 A0A0N1IJC7 J9JQ25 A0A2H8TQ10 A0A1B6LAA0 A0A0L7RDZ6 A0A3L8DW56 E2BTI8 A0A151IXG5 V9IBY7 A0A088AN12 A0A158NYA9 A0A2A3EFI1 K7IQL3 A0A0N0U4B0 A0A0C9Q925 A0A1B6CT80 A0A2R7W8D6 A0A026W4W3 F4WDN7 A0A195FWF6 A0A151I649 A0A151WIL4 A0A154PCZ6 A0A067QZ16 A0A2J7RP32 A0A232ENJ4 A0A2K8JMK5 A0A0T6B9T4 A0A310SUB4 A0A1B6JGX8 E0W0F0 A0A0A9YGG1 A0A0P4VNY1 R4G7R1 A0A1V1G1B5 A0A3B0KN88 A0A1D2NDH1 B4KYE2 A0A1A9YIE2 A0A182FHS8 A0A1W4USG1 A0A1A9UG86 B3NFS1 A0A0J9RRA7 Q2LYL4 B4H638 B4HJI5 A0A1A9ZRX2 A0A0M4ELZ0 A0A0K8VC88 B4PG26 A0A0M3QW31 Q9VSK1 B4IZE3 A0A1J1IAV4 A0A182P3D8 B4LDS4 A0A182VJF9 A0A182Y0T7 A0A182K3P1 B3M5I5 A0A336LWW7 A0A182JGB4 A0A084VDL5 A0A182M715 A0A182UCD6 A0A182XCT5 Q7Q780 A0A182HM62 B4N6D8 A0A182QTC8 A0A034V5E2 A0A182NMA6 Q16G53 A0A182RQA6 A0A1B6GXJ8 A0A0C9RQL4 A0A1I8PZR6 A0A182WK74 W8BGU0 B0WYC2 A0A226F6N1 A0A182HEV5 W5JH31 A0A1S4G226 A0A1A9WVW6
Pubmed
19121390
26354079
28756777
18362917
19820115
28004739
+ More
30249741 20798317 21347285 20075255 24508170 21719571 24845553 28648823 20566863 25401762 27129103 28410430 27289101 17994087 22936249 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 24438588 12364791 25348373 17510324 24495485 26483478 20920257 23761445
30249741 20798317 21347285 20075255 24508170 21719571 24845553 28648823 20566863 25401762 27129103 28410430 27289101 17994087 22936249 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25244985 24438588 12364791 25348373 17510324 24495485 26483478 20920257 23761445
EMBL
BABH01040659
RSAL01000025
RVE52107.1
KQ459460
KPJ00685.1
KZ149937
+ More
PZC77106.1 NWSH01002833 PCG67466.1 ODYU01008607 SOQ52336.1 KQ971354 EFA06028.2 GEZM01067636 JAV67466.1 KQ460009 KPJ18481.1 KC282393 AGX25150.1 KQ459977 KPJ19127.1 ABLF02027290 GFXV01004284 MBW16089.1 GEBQ01019337 JAT20640.1 KQ414613 KOC69044.1 QOIP01000003 RLU24684.1 GL450410 EFN81016.1 KQ980834 KYN12387.1 JR039205 AEY58580.1 ADTU01003593 ADTU01003594 ADTU01003595 KZ288269 PBC30042.1 KQ435824 KOX72078.1 GBYB01010778 JAG80545.1 GEDC01020717 JAS16581.1 KK854310 PTY14475.1 KK107419 EZA51100.1 GL888089 EGI67698.1 KQ981261 KYN44164.1 KQ976399 KYM92674.1 KQ983089 KYQ47666.1 KQ434864 KZC09128.1 KK852816 KDR15788.1 NEVH01002141 PNF42595.1 NNAY01003137 OXU19920.1 KY031153 ATU82904.1 LJIG01002977 KRT83963.1 KQ759870 OAD62389.1 GECU01009205 JAS98501.1 DS235858 EEB19106.1 GBHO01011473 GBHO01011472 JAG32131.1 JAG32132.1 GDKW01002415 JAI54180.1 ACPB03005901 GAHY01002282 JAA75228.1 FX985364 BAX07377.1 OUUW01000009 SPP85278.1 LJIJ01000078 ODN03318.1 CH933809 EDW17721.1 CH954178 EDV50683.1 CM002912 KMY98386.1 CH379069 EAL29848.2 CH479212 EDW33255.1 CH480815 EDW40709.1 CP012525 ALC44572.1 GDHF01033011 GDHF01030485 GDHF01019503 GDHF01015848 JAI19303.1 JAI21829.1 JAI32811.1 JAI36466.1 CM000159 EDW93170.1 ALC43439.1 AE014296 AY058325 AAF50418.2 AAL13554.1 CH916366 EDV96698.1 CVRI01000042 CRK95569.1 CH940647 EDW68947.1 CH902618 EDV39595.2 UFQS01000259 UFQT01000259 SSX02165.1 SSX22542.1 ATLV01011628 KE524705 KFB36059.1 AXCM01013287 AAAB01008960 EAA11963.4 APCN01004366 CH964154 EDW79927.2 AXCN02000936 GAKP01021590 JAC37362.1 CH478329 EAT33218.1 GECZ01018700 GECZ01002630 JAS51069.1 JAS67139.1 GBYB01010775 JAG80542.1 GAMC01008613 JAB97942.1 DS232186 EDS36999.1 LNIX01000001 OXA64516.1 JXUM01132680 KQ567877 KXJ69242.1 ADMH02001519 ETN62220.1
PZC77106.1 NWSH01002833 PCG67466.1 ODYU01008607 SOQ52336.1 KQ971354 EFA06028.2 GEZM01067636 JAV67466.1 KQ460009 KPJ18481.1 KC282393 AGX25150.1 KQ459977 KPJ19127.1 ABLF02027290 GFXV01004284 MBW16089.1 GEBQ01019337 JAT20640.1 KQ414613 KOC69044.1 QOIP01000003 RLU24684.1 GL450410 EFN81016.1 KQ980834 KYN12387.1 JR039205 AEY58580.1 ADTU01003593 ADTU01003594 ADTU01003595 KZ288269 PBC30042.1 KQ435824 KOX72078.1 GBYB01010778 JAG80545.1 GEDC01020717 JAS16581.1 KK854310 PTY14475.1 KK107419 EZA51100.1 GL888089 EGI67698.1 KQ981261 KYN44164.1 KQ976399 KYM92674.1 KQ983089 KYQ47666.1 KQ434864 KZC09128.1 KK852816 KDR15788.1 NEVH01002141 PNF42595.1 NNAY01003137 OXU19920.1 KY031153 ATU82904.1 LJIG01002977 KRT83963.1 KQ759870 OAD62389.1 GECU01009205 JAS98501.1 DS235858 EEB19106.1 GBHO01011473 GBHO01011472 JAG32131.1 JAG32132.1 GDKW01002415 JAI54180.1 ACPB03005901 GAHY01002282 JAA75228.1 FX985364 BAX07377.1 OUUW01000009 SPP85278.1 LJIJ01000078 ODN03318.1 CH933809 EDW17721.1 CH954178 EDV50683.1 CM002912 KMY98386.1 CH379069 EAL29848.2 CH479212 EDW33255.1 CH480815 EDW40709.1 CP012525 ALC44572.1 GDHF01033011 GDHF01030485 GDHF01019503 GDHF01015848 JAI19303.1 JAI21829.1 JAI32811.1 JAI36466.1 CM000159 EDW93170.1 ALC43439.1 AE014296 AY058325 AAF50418.2 AAL13554.1 CH916366 EDV96698.1 CVRI01000042 CRK95569.1 CH940647 EDW68947.1 CH902618 EDV39595.2 UFQS01000259 UFQT01000259 SSX02165.1 SSX22542.1 ATLV01011628 KE524705 KFB36059.1 AXCM01013287 AAAB01008960 EAA11963.4 APCN01004366 CH964154 EDW79927.2 AXCN02000936 GAKP01021590 JAC37362.1 CH478329 EAT33218.1 GECZ01018700 GECZ01002630 JAS51069.1 JAS67139.1 GBYB01010775 JAG80542.1 GAMC01008613 JAB97942.1 DS232186 EDS36999.1 LNIX01000001 OXA64516.1 JXUM01132680 KQ567877 KXJ69242.1 ADMH02001519 ETN62220.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000218220
UP000007266
UP000053240
+ More
UP000007819 UP000053825 UP000279307 UP000008237 UP000078492 UP000005203 UP000005205 UP000242457 UP000002358 UP000053105 UP000053097 UP000007755 UP000078541 UP000078540 UP000075809 UP000076502 UP000027135 UP000235965 UP000215335 UP000009046 UP000015103 UP000268350 UP000094527 UP000009192 UP000092443 UP000069272 UP000192221 UP000078200 UP000008711 UP000001819 UP000008744 UP000001292 UP000092445 UP000092553 UP000002282 UP000000803 UP000001070 UP000183832 UP000075885 UP000008792 UP000075903 UP000076408 UP000075881 UP000007801 UP000075880 UP000030765 UP000075883 UP000075902 UP000076407 UP000007062 UP000075840 UP000007798 UP000075886 UP000075884 UP000008820 UP000075900 UP000095300 UP000075920 UP000002320 UP000198287 UP000069940 UP000249989 UP000000673 UP000091820
UP000007819 UP000053825 UP000279307 UP000008237 UP000078492 UP000005203 UP000005205 UP000242457 UP000002358 UP000053105 UP000053097 UP000007755 UP000078541 UP000078540 UP000075809 UP000076502 UP000027135 UP000235965 UP000215335 UP000009046 UP000015103 UP000268350 UP000094527 UP000009192 UP000092443 UP000069272 UP000192221 UP000078200 UP000008711 UP000001819 UP000008744 UP000001292 UP000092445 UP000092553 UP000002282 UP000000803 UP000001070 UP000183832 UP000075885 UP000008792 UP000075903 UP000076408 UP000075881 UP000007801 UP000075880 UP000030765 UP000075883 UP000075902 UP000076407 UP000007062 UP000075840 UP000007798 UP000075886 UP000075884 UP000008820 UP000075900 UP000095300 UP000075920 UP000002320 UP000198287 UP000069940 UP000249989 UP000000673 UP000091820
SUPFAM
SSF100895
SSF100895
ProteinModelPortal
H9JVU2
A0A3S2PI71
A0A194Q6Q4
A0A2W1BWQ1
A0A2A4J6V9
A0A2H1WHA9
+ More
D6WQS6 A0A1Y1L7A7 A0A0N1IBU2 V9PBG1 A0A0N1IJC7 J9JQ25 A0A2H8TQ10 A0A1B6LAA0 A0A0L7RDZ6 A0A3L8DW56 E2BTI8 A0A151IXG5 V9IBY7 A0A088AN12 A0A158NYA9 A0A2A3EFI1 K7IQL3 A0A0N0U4B0 A0A0C9Q925 A0A1B6CT80 A0A2R7W8D6 A0A026W4W3 F4WDN7 A0A195FWF6 A0A151I649 A0A151WIL4 A0A154PCZ6 A0A067QZ16 A0A2J7RP32 A0A232ENJ4 A0A2K8JMK5 A0A0T6B9T4 A0A310SUB4 A0A1B6JGX8 E0W0F0 A0A0A9YGG1 A0A0P4VNY1 R4G7R1 A0A1V1G1B5 A0A3B0KN88 A0A1D2NDH1 B4KYE2 A0A1A9YIE2 A0A182FHS8 A0A1W4USG1 A0A1A9UG86 B3NFS1 A0A0J9RRA7 Q2LYL4 B4H638 B4HJI5 A0A1A9ZRX2 A0A0M4ELZ0 A0A0K8VC88 B4PG26 A0A0M3QW31 Q9VSK1 B4IZE3 A0A1J1IAV4 A0A182P3D8 B4LDS4 A0A182VJF9 A0A182Y0T7 A0A182K3P1 B3M5I5 A0A336LWW7 A0A182JGB4 A0A084VDL5 A0A182M715 A0A182UCD6 A0A182XCT5 Q7Q780 A0A182HM62 B4N6D8 A0A182QTC8 A0A034V5E2 A0A182NMA6 Q16G53 A0A182RQA6 A0A1B6GXJ8 A0A0C9RQL4 A0A1I8PZR6 A0A182WK74 W8BGU0 B0WYC2 A0A226F6N1 A0A182HEV5 W5JH31 A0A1S4G226 A0A1A9WVW6
D6WQS6 A0A1Y1L7A7 A0A0N1IBU2 V9PBG1 A0A0N1IJC7 J9JQ25 A0A2H8TQ10 A0A1B6LAA0 A0A0L7RDZ6 A0A3L8DW56 E2BTI8 A0A151IXG5 V9IBY7 A0A088AN12 A0A158NYA9 A0A2A3EFI1 K7IQL3 A0A0N0U4B0 A0A0C9Q925 A0A1B6CT80 A0A2R7W8D6 A0A026W4W3 F4WDN7 A0A195FWF6 A0A151I649 A0A151WIL4 A0A154PCZ6 A0A067QZ16 A0A2J7RP32 A0A232ENJ4 A0A2K8JMK5 A0A0T6B9T4 A0A310SUB4 A0A1B6JGX8 E0W0F0 A0A0A9YGG1 A0A0P4VNY1 R4G7R1 A0A1V1G1B5 A0A3B0KN88 A0A1D2NDH1 B4KYE2 A0A1A9YIE2 A0A182FHS8 A0A1W4USG1 A0A1A9UG86 B3NFS1 A0A0J9RRA7 Q2LYL4 B4H638 B4HJI5 A0A1A9ZRX2 A0A0M4ELZ0 A0A0K8VC88 B4PG26 A0A0M3QW31 Q9VSK1 B4IZE3 A0A1J1IAV4 A0A182P3D8 B4LDS4 A0A182VJF9 A0A182Y0T7 A0A182K3P1 B3M5I5 A0A336LWW7 A0A182JGB4 A0A084VDL5 A0A182M715 A0A182UCD6 A0A182XCT5 Q7Q780 A0A182HM62 B4N6D8 A0A182QTC8 A0A034V5E2 A0A182NMA6 Q16G53 A0A182RQA6 A0A1B6GXJ8 A0A0C9RQL4 A0A1I8PZR6 A0A182WK74 W8BGU0 B0WYC2 A0A226F6N1 A0A182HEV5 W5JH31 A0A1S4G226 A0A1A9WVW6
PDB
2P6A
E-value=8.37349e-05,
Score=111
Ontologies
GO
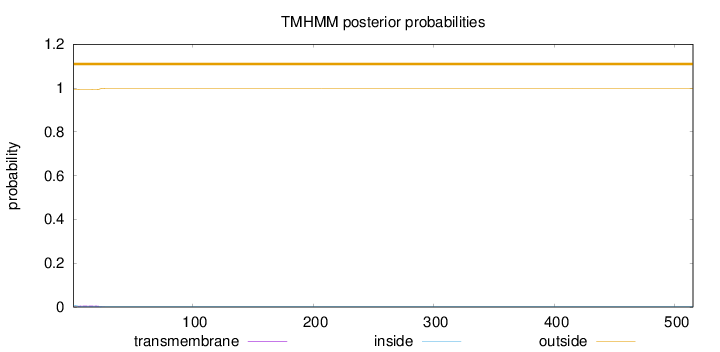
Topology
SignalP
Position: 1 - 22,
Likelihood: 0.999463
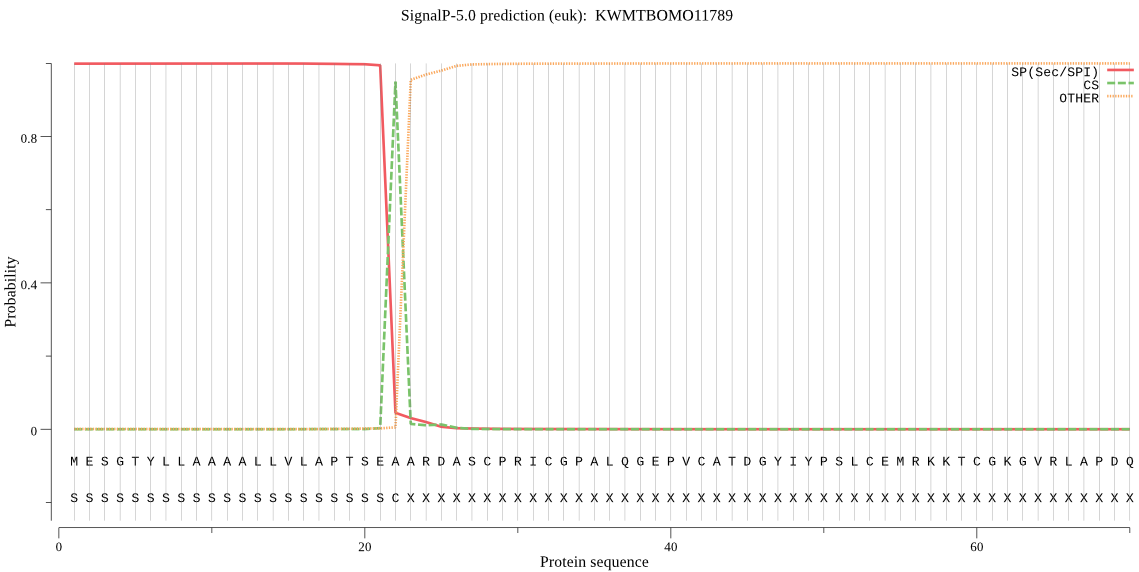
Length:
515
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.1175
Exp number, first 60 AAs:
0.11726
Total prob of N-in:
0.00686
outside
1 - 515
Population Genetic Test Statistics
Pi
261.917232
Theta
208.707431
Tajima's D
1.140228
CLR
0.260488
CSRT
0.697665116744163
Interpretation
Uncertain
