Gene
KWMTBOMO11788
Pre Gene Modal
BGIBMGA013656
Annotation
PREDICTED:_solute_carrier_family_2?_facilitated_glucose_transporter_member_6-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.766
Sequence
CDS
ATGAAGTCCTATTACTTTTTCTTCCGCGAAGGCAGCAAGATCAACCAGATCCTGTGCGCGGTGCTCATAAACCTGCCAGTGTTCGCGTACGGCGCCAGCATCGGCTGGATGTCGCCCATGACCCTGCTGCTGCAGTCCCCGAACTCACCGCGAGACCCGCCCCTCACCGACGGAGAGGTGTCGTGGATGGGTGCGGCCGCGTACCTCACGTGTGTGCCGGCAGACTACTTCATGGCCTTCCTCGGAGACAGGGTCGGCAGGAAGCCGGCGCTTCTGTTCGTCTCCTCCGTTGCTGTCCTGTGCTGGGTGCTGAAGCTGTCGTCTATGGAGACGTGGGCGCTGGTGCTGGCGCGCGCCCTCGTCGGCATCACGATGGCGGGCAGCTACGTGACCTGTCCGCTCTACACCAAAGAGATCAGCGAGGACTCCGTGCGGGGTCTGCTCGGCAGCGTGGTGACATTGTTCCACTGCGCGGGCAACCTGTTCCTGTACGTGGTGGGCGACGTGCTCCCGTACCGCGCCGTGCTGTGGCTGTGTCTGGCGCTGCCCACCCTCCACCTCGTGCTCTTCATGGCCATGCCGGAGTCTCCTTCCTACCTCATTAAGCACGGGAAGCTAGATGAGGCGGTGAGGGTGGTGGCGTGGCTGCGTTGCAGGAGGGAGGACGATCCTCACGTCTTAAATGAGGTTGATCTCATCAAAAACGAGCAGGAGAGGGACCAAGAGAGCAGCAAGTTCGTGCTCAAAGCTGTCGTGACGGACAGGCTGATGAGCCGCGCGTTCCGCATCGCGCTGGCGGTGGCACTGGCGCGCGAGGTGTGCGGCGCCATCCCGGTGCTCAACTTCGCGGGCGAGATCTTCACGCTGGCGGCGGGCGGAGCGCGGCCCCCGCTCTCCCCCAACCAGCAGGCCATGGCGCTGGGCGCCGTGCAGGTCGCCGGCTCCGTGCTCGCCTCCAGCATCGTCGAGAGGGCCGGCAGGAAGCCGCTGCTGCTGACGACGGCGCTGGTGTCGGGCGCCAGCATGTGCGCGCTGGCCACGTGGTTCCTGTGTCGCCGGTGGGGCGCGGAGCTCCCCGCCTCGCTGCCGGTGCTCGCGCTCTGTCTCTGCATCTTCTGTGACGCGGCAGGACTGCAGCCAGTCTCCGTCGTCATCACGGGGGAAATATTCTCATTCAAGTACCGCGGCACGGTGATGGCGACCACCATGGCGGCGTCCTCGTTCGCCGCCTTCCTGCAGATGCTGTTCTTCAAGCCGGTCGCGAACGTGGTGGGACTCTATGCGGCTTTCTACTTCTTCGGAGGCGTGTGCTTACTCACGGCACTCTATGTGGTCCTGGTAGTTCCCGAAACAAAAAAAAGACGAATTGACGAGATCTACGAAGACTTGAAGACTAAAAAAGAGATATTAGAAGAGAAAAAAAATGTAATTGACAAGAAATCTATCCAGGAGTAG
Protein
MKSYYFFFREGSKINQILCAVLINLPVFAYGASIGWMSPMTLLLQSPNSPRDPPLTDGEVSWMGAAAYLTCVPADYFMAFLGDRVGRKPALLFVSSVAVLCWVLKLSSMETWALVLARALVGITMAGSYVTCPLYTKEISEDSVRGLLGSVVTLFHCAGNLFLYVVGDVLPYRAVLWLCLALPTLHLVLFMAMPESPSYLIKHGKLDEAVRVVAWLRCRREDDPHVLNEVDLIKNEQERDQESSKFVLKAVVTDRLMSRAFRIALAVALAREVCGAIPVLNFAGEIFTLAAGGARPPLSPNQQAMALGAVQVAGSVLASSIVERAGRKPLLLTTALVSGASMCALATWFLCRRWGAELPASLPVLALCLCIFCDAAGLQPVSVVITGEIFSFKYRGTVMATTMAASSFAAFLQMLFFKPVANVVGLYAAFYFFGGVCLLTALYVVLVVPETKKRRIDEIYEDLKTKKEILEEKKNVIDKKSIQE
Summary
Similarity
Belongs to the major facilitator superfamily. Sugar transporter (TC 2.A.1.1) family.
Uniprot
H9JVU3
A0A2A4J7G4
A0A194QB83
A0A2W1BQ47
A0A2H1WQ53
A0A2Z5U782
+ More
A0A212FB89 A0A0N1IHX2 A0A212F931 A0A0N1IIG7 A0A2A4JHM3 A0A212FI50 A0A3S2L5Q4 A0A194Q5M1 A0A2H1WE18 A0A2W1BV28 A0A194Q5F6 A0A3S2TMD3 A0A194QQ84 A0A2W1BRA6 A0A3S2TIH7 A0A2A4J8P9 A0A2A4JII3 A0A194Q5B9 A0A212F0G4 A0A212EX46 H9J4J6 A0A2H1WGI4 A0A212FAG2 H9J4P6 A0A0N0PER8 A0A194Q5G2 A0A0N1PHR0 A0A2H1WM77 A0A2A4JKE4 A0A0N0PEG1 A0A2H1WR29 A0A2W1BSA0 A0A194Q5D6 A0A2A4JNL8 A0A2H1X046 A0A2A4J9N1 A0A212EZE7 A0A0N1IJR5 A0A0L7L525 A0A0L7LE34 A0A212F0M2 A0A2J7R8M9 A0A2P8ZDK3 A0A1B6KIF4 A0A2P8ZDL6 A0A1B6CY32 A0A2P8XJ31 A0A1B6JA16 H9JVT8 A0A195FTD3 A0A1B6INA2 A0A1B6IGA4 A0A1B6KWW1 A0A1B6DME3 A0A0C9RQR2 A0A1S3D095 A0A0J7NVW1 A0A3L8DXN6 F4WBY5 A0A2P8ZM01 A0A2P8Z801 A0A0C9RCE9 A0A1B6GYS0 A0A195BM51 A0A158NZ11 A0A336M068 A0A1B6FN92 A0A151J628 A0A154PB56 A0A0A9X9R8 A0A151IEK9 A0A2P8ZLZ8 A0A146L3L7 A0A2H8TZK4 A0A182TXA5 B0WPL1 A0A182UZ66 A0A158NGK6 A0A182VUC8 A0A1B6JU19 A0A182JZA6 A0A195B938 A0A067R5K2 A0A1Q3FRY2 A0A224XJ44 A0A195EU19
A0A212FB89 A0A0N1IHX2 A0A212F931 A0A0N1IIG7 A0A2A4JHM3 A0A212FI50 A0A3S2L5Q4 A0A194Q5M1 A0A2H1WE18 A0A2W1BV28 A0A194Q5F6 A0A3S2TMD3 A0A194QQ84 A0A2W1BRA6 A0A3S2TIH7 A0A2A4J8P9 A0A2A4JII3 A0A194Q5B9 A0A212F0G4 A0A212EX46 H9J4J6 A0A2H1WGI4 A0A212FAG2 H9J4P6 A0A0N0PER8 A0A194Q5G2 A0A0N1PHR0 A0A2H1WM77 A0A2A4JKE4 A0A0N0PEG1 A0A2H1WR29 A0A2W1BSA0 A0A194Q5D6 A0A2A4JNL8 A0A2H1X046 A0A2A4J9N1 A0A212EZE7 A0A0N1IJR5 A0A0L7L525 A0A0L7LE34 A0A212F0M2 A0A2J7R8M9 A0A2P8ZDK3 A0A1B6KIF4 A0A2P8ZDL6 A0A1B6CY32 A0A2P8XJ31 A0A1B6JA16 H9JVT8 A0A195FTD3 A0A1B6INA2 A0A1B6IGA4 A0A1B6KWW1 A0A1B6DME3 A0A0C9RQR2 A0A1S3D095 A0A0J7NVW1 A0A3L8DXN6 F4WBY5 A0A2P8ZM01 A0A2P8Z801 A0A0C9RCE9 A0A1B6GYS0 A0A195BM51 A0A158NZ11 A0A336M068 A0A1B6FN92 A0A151J628 A0A154PB56 A0A0A9X9R8 A0A151IEK9 A0A2P8ZLZ8 A0A146L3L7 A0A2H8TZK4 A0A182TXA5 B0WPL1 A0A182UZ66 A0A158NGK6 A0A182VUC8 A0A1B6JU19 A0A182JZA6 A0A195B938 A0A067R5K2 A0A1Q3FRY2 A0A224XJ44 A0A195EU19
Pubmed
EMBL
BABH01040655
NWSH01002833
PCG67464.1
KQ459460
KPJ00686.1
KZ149937
+ More
PZC77108.1 ODYU01010219 SOQ55189.1 AP017499 BBB06788.1 AGBW02009367 OWR50987.1 KQ460009 KPJ18480.1 AGBW02009650 OWR50256.1 KQ459838 KPJ19742.1 NWSH01001423 PCG71276.1 AGBW02008435 OWR53407.1 RSAL01000132 RVE46342.1 KPJ00847.1 ODYU01008039 SOQ51308.1 KZ149954 PZC76506.1 KPJ00782.1 RSAL01000053 RVE50024.1 KQ461181 KPJ07673.1 KZ149924 PZC77632.1 RSAL01000114 RVE46930.1 NWSH01002620 PCG67830.1 NWSH01001270 PCG71905.1 KPJ00738.1 AGBW02011078 OWR47246.1 AGBW02011864 OWR46004.1 BABH01021836 ODYU01008186 SOQ51574.1 AGBW02009482 OWR50710.1 BABH01022059 KPJ19739.1 KPJ00783.1 KQ460654 KPJ12970.1 ODYU01009532 SOQ54032.1 PCG71903.1 KPJ19738.1 ODYU01010429 SOQ55525.1 PZC76505.1 KPJ00762.1 NWSH01001033 PCG73010.1 ODYU01012350 SOQ58622.1 NWSH01002244 PCG68837.1 AGBW02011317 OWR46831.1 KPJ19754.1 JTDY01002946 KOB70431.1 JTDY01001495 KOB73737.1 AGBW02011050 OWR47308.1 NEVH01006721 PNF37175.1 PYGN01000088 PSN54587.1 GEBQ01028740 JAT11237.1 PSN54585.1 GEDC01018964 JAS18334.1 PYGN01001954 PSN32011.1 GECU01011708 JAS95998.1 BABH01040665 BABH01040666 BABH01040667 KQ981276 KYN43567.1 GECU01019308 JAS88398.1 GECU01021763 JAS85943.1 GEBQ01024034 GEBQ01002289 JAT15943.1 JAT37688.1 GEDC01010470 JAS26828.1 GBYB01009586 JAG79353.1 LBMM01001313 KMQ96530.1 QOIP01000003 RLU25240.1 GL888067 EGI68274.1 PYGN01000019 PSN57531.1 PYGN01000154 PSN52620.1 GBYB01005985 JAG75752.1 GECZ01003576 GECZ01002212 JAS66193.1 JAS67557.1 KQ976444 KYM86198.1 ADTU01004466 UFQT01000067 SSX19358.1 GECZ01018093 JAS51676.1 KQ979883 KYN18677.1 KQ434864 KZC09071.1 GBHO01026885 GBRD01008360 JAG16719.1 JAG57461.1 KQ977889 KYM99007.1 PSN57528.1 GDHC01015631 JAQ02998.1 GFXV01007889 MBW19694.1 DS232026 EDS32412.1 ADTU01015114 GECU01005303 JAT02404.1 KQ976542 KYM81051.1 KK852870 KDR14606.1 GFDL01004852 JAV30193.1 GFTR01006588 JAW09838.1 KQ981965 KYN31745.1
PZC77108.1 ODYU01010219 SOQ55189.1 AP017499 BBB06788.1 AGBW02009367 OWR50987.1 KQ460009 KPJ18480.1 AGBW02009650 OWR50256.1 KQ459838 KPJ19742.1 NWSH01001423 PCG71276.1 AGBW02008435 OWR53407.1 RSAL01000132 RVE46342.1 KPJ00847.1 ODYU01008039 SOQ51308.1 KZ149954 PZC76506.1 KPJ00782.1 RSAL01000053 RVE50024.1 KQ461181 KPJ07673.1 KZ149924 PZC77632.1 RSAL01000114 RVE46930.1 NWSH01002620 PCG67830.1 NWSH01001270 PCG71905.1 KPJ00738.1 AGBW02011078 OWR47246.1 AGBW02011864 OWR46004.1 BABH01021836 ODYU01008186 SOQ51574.1 AGBW02009482 OWR50710.1 BABH01022059 KPJ19739.1 KPJ00783.1 KQ460654 KPJ12970.1 ODYU01009532 SOQ54032.1 PCG71903.1 KPJ19738.1 ODYU01010429 SOQ55525.1 PZC76505.1 KPJ00762.1 NWSH01001033 PCG73010.1 ODYU01012350 SOQ58622.1 NWSH01002244 PCG68837.1 AGBW02011317 OWR46831.1 KPJ19754.1 JTDY01002946 KOB70431.1 JTDY01001495 KOB73737.1 AGBW02011050 OWR47308.1 NEVH01006721 PNF37175.1 PYGN01000088 PSN54587.1 GEBQ01028740 JAT11237.1 PSN54585.1 GEDC01018964 JAS18334.1 PYGN01001954 PSN32011.1 GECU01011708 JAS95998.1 BABH01040665 BABH01040666 BABH01040667 KQ981276 KYN43567.1 GECU01019308 JAS88398.1 GECU01021763 JAS85943.1 GEBQ01024034 GEBQ01002289 JAT15943.1 JAT37688.1 GEDC01010470 JAS26828.1 GBYB01009586 JAG79353.1 LBMM01001313 KMQ96530.1 QOIP01000003 RLU25240.1 GL888067 EGI68274.1 PYGN01000019 PSN57531.1 PYGN01000154 PSN52620.1 GBYB01005985 JAG75752.1 GECZ01003576 GECZ01002212 JAS66193.1 JAS67557.1 KQ976444 KYM86198.1 ADTU01004466 UFQT01000067 SSX19358.1 GECZ01018093 JAS51676.1 KQ979883 KYN18677.1 KQ434864 KZC09071.1 GBHO01026885 GBRD01008360 JAG16719.1 JAG57461.1 KQ977889 KYM99007.1 PSN57528.1 GDHC01015631 JAQ02998.1 GFXV01007889 MBW19694.1 DS232026 EDS32412.1 ADTU01015114 GECU01005303 JAT02404.1 KQ976542 KYM81051.1 KK852870 KDR14606.1 GFDL01004852 JAV30193.1 GFTR01006588 JAW09838.1 KQ981965 KYN31745.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000007151
UP000053240
UP000283053
+ More
UP000037510 UP000235965 UP000245037 UP000078541 UP000079169 UP000036403 UP000279307 UP000007755 UP000078540 UP000005205 UP000078492 UP000076502 UP000078542 UP000075902 UP000002320 UP000075903 UP000075920 UP000075881 UP000027135
UP000037510 UP000235965 UP000245037 UP000078541 UP000079169 UP000036403 UP000279307 UP000007755 UP000078540 UP000005205 UP000078492 UP000076502 UP000078542 UP000075902 UP000002320 UP000075903 UP000075920 UP000075881 UP000027135
Interpro
CDD
ProteinModelPortal
H9JVU3
A0A2A4J7G4
A0A194QB83
A0A2W1BQ47
A0A2H1WQ53
A0A2Z5U782
+ More
A0A212FB89 A0A0N1IHX2 A0A212F931 A0A0N1IIG7 A0A2A4JHM3 A0A212FI50 A0A3S2L5Q4 A0A194Q5M1 A0A2H1WE18 A0A2W1BV28 A0A194Q5F6 A0A3S2TMD3 A0A194QQ84 A0A2W1BRA6 A0A3S2TIH7 A0A2A4J8P9 A0A2A4JII3 A0A194Q5B9 A0A212F0G4 A0A212EX46 H9J4J6 A0A2H1WGI4 A0A212FAG2 H9J4P6 A0A0N0PER8 A0A194Q5G2 A0A0N1PHR0 A0A2H1WM77 A0A2A4JKE4 A0A0N0PEG1 A0A2H1WR29 A0A2W1BSA0 A0A194Q5D6 A0A2A4JNL8 A0A2H1X046 A0A2A4J9N1 A0A212EZE7 A0A0N1IJR5 A0A0L7L525 A0A0L7LE34 A0A212F0M2 A0A2J7R8M9 A0A2P8ZDK3 A0A1B6KIF4 A0A2P8ZDL6 A0A1B6CY32 A0A2P8XJ31 A0A1B6JA16 H9JVT8 A0A195FTD3 A0A1B6INA2 A0A1B6IGA4 A0A1B6KWW1 A0A1B6DME3 A0A0C9RQR2 A0A1S3D095 A0A0J7NVW1 A0A3L8DXN6 F4WBY5 A0A2P8ZM01 A0A2P8Z801 A0A0C9RCE9 A0A1B6GYS0 A0A195BM51 A0A158NZ11 A0A336M068 A0A1B6FN92 A0A151J628 A0A154PB56 A0A0A9X9R8 A0A151IEK9 A0A2P8ZLZ8 A0A146L3L7 A0A2H8TZK4 A0A182TXA5 B0WPL1 A0A182UZ66 A0A158NGK6 A0A182VUC8 A0A1B6JU19 A0A182JZA6 A0A195B938 A0A067R5K2 A0A1Q3FRY2 A0A224XJ44 A0A195EU19
A0A212FB89 A0A0N1IHX2 A0A212F931 A0A0N1IIG7 A0A2A4JHM3 A0A212FI50 A0A3S2L5Q4 A0A194Q5M1 A0A2H1WE18 A0A2W1BV28 A0A194Q5F6 A0A3S2TMD3 A0A194QQ84 A0A2W1BRA6 A0A3S2TIH7 A0A2A4J8P9 A0A2A4JII3 A0A194Q5B9 A0A212F0G4 A0A212EX46 H9J4J6 A0A2H1WGI4 A0A212FAG2 H9J4P6 A0A0N0PER8 A0A194Q5G2 A0A0N1PHR0 A0A2H1WM77 A0A2A4JKE4 A0A0N0PEG1 A0A2H1WR29 A0A2W1BSA0 A0A194Q5D6 A0A2A4JNL8 A0A2H1X046 A0A2A4J9N1 A0A212EZE7 A0A0N1IJR5 A0A0L7L525 A0A0L7LE34 A0A212F0M2 A0A2J7R8M9 A0A2P8ZDK3 A0A1B6KIF4 A0A2P8ZDL6 A0A1B6CY32 A0A2P8XJ31 A0A1B6JA16 H9JVT8 A0A195FTD3 A0A1B6INA2 A0A1B6IGA4 A0A1B6KWW1 A0A1B6DME3 A0A0C9RQR2 A0A1S3D095 A0A0J7NVW1 A0A3L8DXN6 F4WBY5 A0A2P8ZM01 A0A2P8Z801 A0A0C9RCE9 A0A1B6GYS0 A0A195BM51 A0A158NZ11 A0A336M068 A0A1B6FN92 A0A151J628 A0A154PB56 A0A0A9X9R8 A0A151IEK9 A0A2P8ZLZ8 A0A146L3L7 A0A2H8TZK4 A0A182TXA5 B0WPL1 A0A182UZ66 A0A158NGK6 A0A182VUC8 A0A1B6JU19 A0A182JZA6 A0A195B938 A0A067R5K2 A0A1Q3FRY2 A0A224XJ44 A0A195EU19
PDB
4LDS
E-value=1.45207e-21,
Score=255
Ontologies
GO
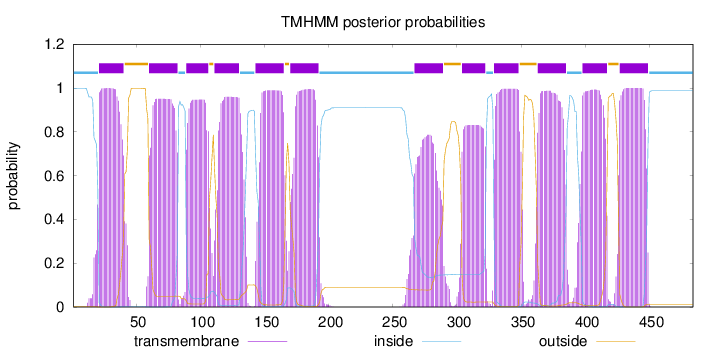
Topology
Length:
484
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
245.82216
Exp number, first 60 AAs:
22.47997
Total prob of N-in:
0.99891
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 40
outside
41 - 59
TMhelix
60 - 82
inside
83 - 88
TMhelix
89 - 106
outside
107 - 110
TMhelix
111 - 130
inside
131 - 142
TMhelix
143 - 165
outside
166 - 169
TMhelix
170 - 192
inside
193 - 266
TMhelix
267 - 289
outside
290 - 303
TMhelix
304 - 322
inside
323 - 328
TMhelix
329 - 348
outside
349 - 362
TMhelix
363 - 385
inside
386 - 397
TMhelix
398 - 417
outside
418 - 426
TMhelix
427 - 449
inside
450 - 484
Population Genetic Test Statistics
Pi
234.799002
Theta
175.331765
Tajima's D
1.257759
CLR
0.352658
CSRT
0.729163541822909
Interpretation
Uncertain
