Gene
KWMTBOMO11787
Annotation
gustatory_receptor_60_[Bombyx_mori]
Full name
Gustatory receptor
Location in the cell
PlasmaMembrane Reliability : 4.621
Sequence
CDS
ATGTTGACACCACGCAGCGACCTTTGCAACGAAAAACTTTCACCCAGCTTTCCATCTGGAAAGACGACAGCTGCAGATAAGGACGACACGGAGGCGCGGTGCCAGGTGGACAGCAGCCTCGAGCGGCTGCTGCTTCCGTTCAACTTGGTGCAGCACGTGTCGTTCATACCGATGTACAGCATCCGCCGCGGGCTGGTCTCGCCCGACGGACCGCTCGCCTACCTCTACTCGCTCCTCGGCTTCTGTTTGTTCACGTCCGTCTCCGTCTACCGCAACGCGATCATGCACGGGACGCGGCTCTCCTCCCTCCACCTCTTCACCCTGTACTCCGACTTGGTCTCGTTCGTGATCAACTACTCGCTCAGCTTGATCTGCAACGTGGTCAACAGCAAGAGCAACGTGGAGTTCGTGTGTCGGCTGCAGCGGCTGCAGACCGTCCTGCGACGCAACCAGCGGGAGCAAGAGCAGTTCGCTCGGAGCAACTGGGCGCACCTGGCGGTGGTGACGGCCCTGTACCTCGCCGTCGTGGGGCTGCTCAACGTGGTGGTGCTGAAGCAGTCGTTGCCCGACACCCTGTACTTGTTGCTGCTCTTCTGCATCGACGTGAACGTACTTTACGCGACGAGGATGCTGGCCCTGCTCCGGTGCTACCTTCAGCTCTGGACGCGTAAAATTAACGAGAAGGCGTTCAATCCAGTGCACCACAACATGTTCACGGCTTATTTGGATATTTTGCAAGAGTACGAGGTCTACACAACTCTCTTTAAAAAAATTATCACGTACTATGTATTGGAAACGTTCTTGCACGGCCTGCTGTACGTACAGGTCGCGATACAGATCTGCAAGTCCATCCGTAGATCCGGACGTTTTAGCGAGCAGCTGATGATGATCGTTTCAATCTTCACGTGGACGATCAAGAACATGATAATAATGACGCTTCACAACGTGGAGTGCGAAAAGTTCTACCTCGCGGTGGAGCAGGCCGTCGCCGCCTGCCAGACGCAGCGCGCCTCCACCACGCGATGCCGTAACTAA
Protein
MLTPRSDLCNEKLSPSFPSGKTTAADKDDTEARCQVDSSLERLLLPFNLVQHVSFIPMYSIRRGLVSPDGPLAYLYSLLGFCLFTSVSVYRNAIMHGTRLSSLHLFTLYSDLVSFVINYSLSLICNVVNSKSNVEFVCRLQRLQTVLRRNQREQEQFARSNWAHLAVVTALYLAVVGLLNVVVLKQSLPDTLYLLLLFCIDVNVLYATRMLALLRCYLQLWTRKINEKAFNPVHHNMFTAYLDILQEYEVYTTLFKKIITYYVLETFLHGLLYVQVAIQICKSIRRSGRFSEQLMMIVSIFTWTIKNMIIMTLHNVECEKFYLAVEQAVAACQTQRASTTRCRN
Summary
Description
Gustatory receptor which mediates acceptance or avoidance behavior, depending on its substrates.
Similarity
Belongs to the insect chemoreceptor superfamily. Gustatory receptor (GR) family.
Uniprot
B3GTE1
A0A2W1BKG8
A0A2H1W0P7
A0A2H1VPC4
A0A2H1WAS7
A0A2W1BTU2
+ More
A0A2H1W8E9 A0A2H1WXE2 A0A2H1V7B1 A0A2H1WMK8 A0A2H1W7R4 A0A2H1WBX4 A0A2H1VR14 A0A2H1VPB9 A0A2H1WDS9 A0A2H1WC59 A0A2H1VPB7 A0A2H1W875 B7FF45 A0A2H1WBV5 A0A2H1W7Z4 A0A2H1VSY7 A0A2H1VUK0 A0A2H1V7D3 A0A2H1V7B8 A0A2H1W9R2 A0A2H1WDS3 A0A2H1WYK5 A0A2H1VCJ8 A0A2H1W8T8 A0A2H1W7Q9 A0A2W1BI01 A0A2H1WCJ3 A0A2H1WX57 A0A2H1WVG9 A0A2H1X4N6 A0A2H1WNG2 A0A2H1VTH0 A0A2H1W5P4 A0A2H1V8I5 A0A2H1VPG1 A0A2W1BPU3 A0A2W1BI00 A0A223HCW3 A0A2H1V7C5 A0A2H1W9V0 A0A2W1BWJ2 A0A2H1V7E3 A0A2H1VPF1 A0A2H1V7D5 A0A2H1V7G9 A0A2H1W3G7 A0A2H1WBG5 A0A2H1WV94 A0A2H1V7C3 A0A2H1VKM5 A0A2H1WDR7 A0A2H1VUI2 A0A2W1BTV4 A0A2H1WBG4 A0A2H1WEB1 A0A2H1WAT0 A0A2H1V907 A0A2H1VJI5 A0A2H1WE39 A0A2H1V8J8 A0A2H1V8L7 A0A2H1V7C1 A0A2H1VPD4 A0A2H1W840 A0A2H1V7F6 A0A2H1WX43 A0A2H1WBW4 A0A2H1VJH3 A0A2H1V7G3 A0A2H1VSZ9 A0A2W1BQ12 A0A2H1V7A2 A0A2H1X289 A0A223HCW2 A0A2H1VT21 A0A2H1V7F3 A0A2H1WDS7 A0A2H1VJI8 A0A2H1VPC7 A0A2H1W859 A0A2H1V7E0 A0A2A4K7F9 A0A2H1WQB3 A0A2H1VPF8 A0A2H1VZF7 A0A2H1W871
A0A2H1W8E9 A0A2H1WXE2 A0A2H1V7B1 A0A2H1WMK8 A0A2H1W7R4 A0A2H1WBX4 A0A2H1VR14 A0A2H1VPB9 A0A2H1WDS9 A0A2H1WC59 A0A2H1VPB7 A0A2H1W875 B7FF45 A0A2H1WBV5 A0A2H1W7Z4 A0A2H1VSY7 A0A2H1VUK0 A0A2H1V7D3 A0A2H1V7B8 A0A2H1W9R2 A0A2H1WDS3 A0A2H1WYK5 A0A2H1VCJ8 A0A2H1W8T8 A0A2H1W7Q9 A0A2W1BI01 A0A2H1WCJ3 A0A2H1WX57 A0A2H1WVG9 A0A2H1X4N6 A0A2H1WNG2 A0A2H1VTH0 A0A2H1W5P4 A0A2H1V8I5 A0A2H1VPG1 A0A2W1BPU3 A0A2W1BI00 A0A223HCW3 A0A2H1V7C5 A0A2H1W9V0 A0A2W1BWJ2 A0A2H1V7E3 A0A2H1VPF1 A0A2H1V7D5 A0A2H1V7G9 A0A2H1W3G7 A0A2H1WBG5 A0A2H1WV94 A0A2H1V7C3 A0A2H1VKM5 A0A2H1WDR7 A0A2H1VUI2 A0A2W1BTV4 A0A2H1WBG4 A0A2H1WEB1 A0A2H1WAT0 A0A2H1V907 A0A2H1VJI5 A0A2H1WE39 A0A2H1V8J8 A0A2H1V8L7 A0A2H1V7C1 A0A2H1VPD4 A0A2H1W840 A0A2H1V7F6 A0A2H1WX43 A0A2H1WBW4 A0A2H1VJH3 A0A2H1V7G3 A0A2H1VSZ9 A0A2W1BQ12 A0A2H1V7A2 A0A2H1X289 A0A223HCW2 A0A2H1VT21 A0A2H1V7F3 A0A2H1WDS7 A0A2H1VJI8 A0A2H1VPC7 A0A2H1W859 A0A2H1V7E0 A0A2A4K7F9 A0A2H1WQB3 A0A2H1VPF8 A0A2H1VZF7 A0A2H1W871
EMBL
EU769124
ACD85127.1
KZ150111
PZC73336.1
ODYU01005551
SOQ46506.1
+ More
ODYU01003652 SOQ42680.1 ODYU01007408 SOQ50147.1 KZ149937 PZC77065.1 ODYU01006930 SOQ49216.1 ODYU01011791 SOQ57739.1 ODYU01001051 SOQ36735.1 ODYU01009674 SOQ54268.1 ODYU01006874 SOQ49110.1 ODYU01007624 SOQ50560.1 SOQ42684.1 SOQ42681.1 ODYU01007981 SOQ51219.1 ODYU01007552 SOQ50422.1 SOQ42679.1 SOQ49213.1 BK006612 DAA06392.1 SOQ50561.1 SOQ49215.1 ODYU01004258 SOQ43937.1 ODYU01004526 SOQ44500.1 SOQ36731.1 SOQ36745.1 SOQ49214.1 SOQ51218.1 ODYU01012012 SOQ58082.1 ODYU01001808 SOQ38516.1 ODYU01006944 SOQ49252.1 SOQ49111.1 PZC73334.1 SOQ50562.1 ODYU01011664 SOQ57556.1 ODYU01011315 SOQ56982.1 ODYU01013004 SOQ59604.1 ODYU01009893 SOQ54613.1 ODYU01004314 SOQ44056.1 ODYU01006369 SOQ48162.1 SOQ36722.1 SOQ42687.1 PZC77062.1 PZC73335.1 KY283557 AST36217.1 SOQ36750.1 SOQ49254.1 PZC77046.1 SOQ36719.1 SOQ42677.1 SOQ36748.1 SOQ36757.1 ODYU01006103 SOQ47649.1 ODYU01007551 SOQ50419.1 SOQ56983.1 SOQ36721.1 ODYU01002909 SOQ40992.1 SOQ51209.1 SOQ44499.1 PZC77064.1 SOQ50421.1 ODYU01008107 SOQ51420.1 ODYU01007110 SOQ49594.1 SOQ36734.1 SOQ40990.1 SOQ51216.1 SOQ36732.1 SOQ36752.1 SOQ36743.1 SOQ42690.1 SOQ49217.1 SOQ36751.1 SOQ57557.1 SOQ50558.1 SOQ40989.1 SOQ36739.1 SOQ43939.1 PZC77068.1 SOQ36730.1 ODYU01012828 SOQ59326.1 KY283553 KY283556 AST36216.1 SOQ43938.1 SOQ36729.1 SOQ51213.1 SOQ40988.1 SOQ42685.1 SOQ49248.1 SOQ36727.1 NWSH01000066 PCG80009.1 ODYU01010212 SOQ55176.1 SOQ42686.1 ODYU01005405 SOQ46208.1 SOQ49247.1
ODYU01003652 SOQ42680.1 ODYU01007408 SOQ50147.1 KZ149937 PZC77065.1 ODYU01006930 SOQ49216.1 ODYU01011791 SOQ57739.1 ODYU01001051 SOQ36735.1 ODYU01009674 SOQ54268.1 ODYU01006874 SOQ49110.1 ODYU01007624 SOQ50560.1 SOQ42684.1 SOQ42681.1 ODYU01007981 SOQ51219.1 ODYU01007552 SOQ50422.1 SOQ42679.1 SOQ49213.1 BK006612 DAA06392.1 SOQ50561.1 SOQ49215.1 ODYU01004258 SOQ43937.1 ODYU01004526 SOQ44500.1 SOQ36731.1 SOQ36745.1 SOQ49214.1 SOQ51218.1 ODYU01012012 SOQ58082.1 ODYU01001808 SOQ38516.1 ODYU01006944 SOQ49252.1 SOQ49111.1 PZC73334.1 SOQ50562.1 ODYU01011664 SOQ57556.1 ODYU01011315 SOQ56982.1 ODYU01013004 SOQ59604.1 ODYU01009893 SOQ54613.1 ODYU01004314 SOQ44056.1 ODYU01006369 SOQ48162.1 SOQ36722.1 SOQ42687.1 PZC77062.1 PZC73335.1 KY283557 AST36217.1 SOQ36750.1 SOQ49254.1 PZC77046.1 SOQ36719.1 SOQ42677.1 SOQ36748.1 SOQ36757.1 ODYU01006103 SOQ47649.1 ODYU01007551 SOQ50419.1 SOQ56983.1 SOQ36721.1 ODYU01002909 SOQ40992.1 SOQ51209.1 SOQ44499.1 PZC77064.1 SOQ50421.1 ODYU01008107 SOQ51420.1 ODYU01007110 SOQ49594.1 SOQ36734.1 SOQ40990.1 SOQ51216.1 SOQ36732.1 SOQ36752.1 SOQ36743.1 SOQ42690.1 SOQ49217.1 SOQ36751.1 SOQ57557.1 SOQ50558.1 SOQ40989.1 SOQ36739.1 SOQ43939.1 PZC77068.1 SOQ36730.1 ODYU01012828 SOQ59326.1 KY283553 KY283556 AST36216.1 SOQ43938.1 SOQ36729.1 SOQ51213.1 SOQ40988.1 SOQ42685.1 SOQ49248.1 SOQ36727.1 NWSH01000066 PCG80009.1 ODYU01010212 SOQ55176.1 SOQ42686.1 ODYU01005405 SOQ46208.1 SOQ49247.1
Proteomes
Pfam
PF08395 7tm_7
Interpro
IPR013604
7TM_chemorcpt
ProteinModelPortal
B3GTE1
A0A2W1BKG8
A0A2H1W0P7
A0A2H1VPC4
A0A2H1WAS7
A0A2W1BTU2
+ More
A0A2H1W8E9 A0A2H1WXE2 A0A2H1V7B1 A0A2H1WMK8 A0A2H1W7R4 A0A2H1WBX4 A0A2H1VR14 A0A2H1VPB9 A0A2H1WDS9 A0A2H1WC59 A0A2H1VPB7 A0A2H1W875 B7FF45 A0A2H1WBV5 A0A2H1W7Z4 A0A2H1VSY7 A0A2H1VUK0 A0A2H1V7D3 A0A2H1V7B8 A0A2H1W9R2 A0A2H1WDS3 A0A2H1WYK5 A0A2H1VCJ8 A0A2H1W8T8 A0A2H1W7Q9 A0A2W1BI01 A0A2H1WCJ3 A0A2H1WX57 A0A2H1WVG9 A0A2H1X4N6 A0A2H1WNG2 A0A2H1VTH0 A0A2H1W5P4 A0A2H1V8I5 A0A2H1VPG1 A0A2W1BPU3 A0A2W1BI00 A0A223HCW3 A0A2H1V7C5 A0A2H1W9V0 A0A2W1BWJ2 A0A2H1V7E3 A0A2H1VPF1 A0A2H1V7D5 A0A2H1V7G9 A0A2H1W3G7 A0A2H1WBG5 A0A2H1WV94 A0A2H1V7C3 A0A2H1VKM5 A0A2H1WDR7 A0A2H1VUI2 A0A2W1BTV4 A0A2H1WBG4 A0A2H1WEB1 A0A2H1WAT0 A0A2H1V907 A0A2H1VJI5 A0A2H1WE39 A0A2H1V8J8 A0A2H1V8L7 A0A2H1V7C1 A0A2H1VPD4 A0A2H1W840 A0A2H1V7F6 A0A2H1WX43 A0A2H1WBW4 A0A2H1VJH3 A0A2H1V7G3 A0A2H1VSZ9 A0A2W1BQ12 A0A2H1V7A2 A0A2H1X289 A0A223HCW2 A0A2H1VT21 A0A2H1V7F3 A0A2H1WDS7 A0A2H1VJI8 A0A2H1VPC7 A0A2H1W859 A0A2H1V7E0 A0A2A4K7F9 A0A2H1WQB3 A0A2H1VPF8 A0A2H1VZF7 A0A2H1W871
A0A2H1W8E9 A0A2H1WXE2 A0A2H1V7B1 A0A2H1WMK8 A0A2H1W7R4 A0A2H1WBX4 A0A2H1VR14 A0A2H1VPB9 A0A2H1WDS9 A0A2H1WC59 A0A2H1VPB7 A0A2H1W875 B7FF45 A0A2H1WBV5 A0A2H1W7Z4 A0A2H1VSY7 A0A2H1VUK0 A0A2H1V7D3 A0A2H1V7B8 A0A2H1W9R2 A0A2H1WDS3 A0A2H1WYK5 A0A2H1VCJ8 A0A2H1W8T8 A0A2H1W7Q9 A0A2W1BI01 A0A2H1WCJ3 A0A2H1WX57 A0A2H1WVG9 A0A2H1X4N6 A0A2H1WNG2 A0A2H1VTH0 A0A2H1W5P4 A0A2H1V8I5 A0A2H1VPG1 A0A2W1BPU3 A0A2W1BI00 A0A223HCW3 A0A2H1V7C5 A0A2H1W9V0 A0A2W1BWJ2 A0A2H1V7E3 A0A2H1VPF1 A0A2H1V7D5 A0A2H1V7G9 A0A2H1W3G7 A0A2H1WBG5 A0A2H1WV94 A0A2H1V7C3 A0A2H1VKM5 A0A2H1WDR7 A0A2H1VUI2 A0A2W1BTV4 A0A2H1WBG4 A0A2H1WEB1 A0A2H1WAT0 A0A2H1V907 A0A2H1VJI5 A0A2H1WE39 A0A2H1V8J8 A0A2H1V8L7 A0A2H1V7C1 A0A2H1VPD4 A0A2H1W840 A0A2H1V7F6 A0A2H1WX43 A0A2H1WBW4 A0A2H1VJH3 A0A2H1V7G3 A0A2H1VSZ9 A0A2W1BQ12 A0A2H1V7A2 A0A2H1X289 A0A223HCW2 A0A2H1VT21 A0A2H1V7F3 A0A2H1WDS7 A0A2H1VJI8 A0A2H1VPC7 A0A2H1W859 A0A2H1V7E0 A0A2A4K7F9 A0A2H1WQB3 A0A2H1VPF8 A0A2H1VZF7 A0A2H1W871
Ontologies
PANTHER
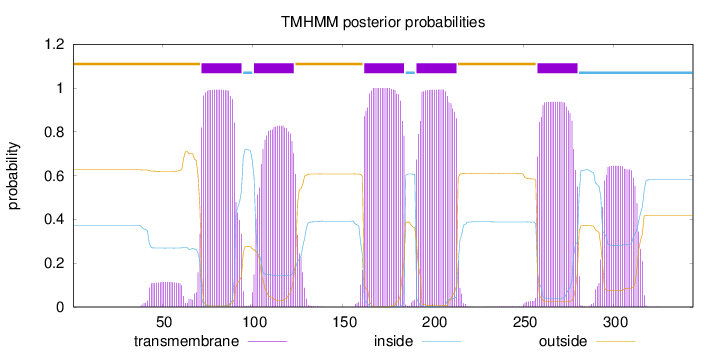
Topology
Subcellular location
Cell membrane
Length:
344
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
120.05509
Exp number, first 60 AAs:
2.061
Total prob of N-in:
0.37354
outside
1 - 71
TMhelix
72 - 94
inside
95 - 100
TMhelix
101 - 123
outside
124 - 161
TMhelix
162 - 184
inside
185 - 190
TMhelix
191 - 213
outside
214 - 257
TMhelix
258 - 280
inside
281 - 344
Population Genetic Test Statistics
Pi
31.017887
Theta
65.764547
Tajima's D
-1.649862
CLR
433.667395
CSRT
0.0412979351032448
Interpretation
Uncertain
