Gene
KWMTBOMO11780 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA014483
Annotation
hypothetical_protein_KGM_02094_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 2.153
Sequence
CDS
ATGTCCTTCTTCTTAAAACGCACTATTGGTGCGGGAAACGGCGTCGGACGAGCTATTTTAAACAGTACCAAAATACCTGGTGTCCAGCGCGTAGCGACAAAATCTACCGAAAAAACCCCAGTACAAGAGACACGACCAACTGTAGCCAAATTAGATCCACTGCAAAAGGCGCAGCTCGTCGACTTCGGCAAATATGTAGCTGACTGTCTACCCAAGTATGTGCAGAAAGTACAGCTGACTGCCGGCAACGAACTAGAGGTGCTCATCCCTCCGGACGCAGTCATCCCGGTTTTATCCTTTTTGAAGGACCACCACAGCGCTCAGTTTGCAAACCTTGTCGATATCGCTGGCATGGACGTACCCTCGCGCTCAAACAGATTCGAAATCATATATAACATACTTTCTCTACGTTACAATGCCCGAATAAGAGTCAAAACTTACACTGACGAGTTGACTCCTGTAGACTCAGCCTGCGATGTATTCAAGGCCGCAAATTGGTACGAACGAGAGATCTGGGACATGTATGGAGTATTTTTTGCTAACCATCCTGATTTAAGAAGGATTTTAACAGATTACGGTTTTGAGGGGCACCCTTTCAGGAAGGACTTCCCTCTAAGTGGATATGTAGAAGTTCGTTATGACGATGAGCAGAAGCGTGTGGTGGTCGAACCCCTTGAGTTGGCACAAGAGTTCAGACGCTTTGAACTGAGTGCACCATGGGAGCAGTTCCCTAACTTTAGAGCCAACCCTGTAACAGAGGAAGTAGTGAACCCGTCTGAGGATCAACCAAAAAAATAA
Protein
MSFFLKRTIGAGNGVGRAILNSTKIPGVQRVATKSTEKTPVQETRPTVAKLDPLQKAQLVDFGKYVADCLPKYVQKVQLTAGNELEVLIPPDAVIPVLSFLKDHHSAQFANLVDIAGMDVPSRSNRFEIIYNILSLRYNARIRVKTYTDELTPVDSACDVFKAANWYEREIWDMYGVFFANHPDLRRILTDYGFEGHPFRKDFPLSGYVEVRYDDEQKRVVVEPLELAQEFRRFELSAPWEQFPNFRANPVTEEVVNPSEDQPKK
Summary
Similarity
Belongs to the complex I 30 kDa subunit family.
Uniprot
H9JY65
A0A212EJY1
A0A2A4JLG1
A0A0L7LIY8
A0A2W1BUD9
A0A2H1WEN3
+ More
I4DQE8 A0A194QXW1 A0A2P8Z6P2 A0A1L8E0I2 U5ETX7 A0A2J7Q2L7 A0A1L8E0N6 A0A1B6JRC9 A0A1B0CFY2 A0A182WC31 A0A1B6ERE2 V5IA77 A0A336LKT7 A0A182T7K3 A0A2M4BXB1 A0A182KBE8 A0A182Q115 T1DER2 T1E7A3 A0A182V682 A0A182U1T4 A0A182X904 A0A182LKX4 Q7Q5G0 A0A182I442 A0A1J1IJC4 A0A1B6CKI0 W5JSE8 A0A2M4BX44 A0A182FF92 A0A2M3Z4C6 A0A182IUK7 A0A2M4AAH3 A0A084VAX3 A0A182LV84 A0A182H793 A0A182PB87 A0A182YNV4 D6WIB0 A0A023ENH9 A0A182NJ35 A0A023EME8 A0A067QGG8 A0A182RMP5 Q29E23 B4H416 B0X266 J3JV29 B4HTK8 B4QP26 A0A3B0JZI3 N6TWH4 B4N6C3 A0A1W4W036 A0A0M4EBY7 A0A1A9WLU0 Q9VZU4 A0A1D2N063 B3M7D6 T1PFE4 B4LFE3 B3NF35 A0A3B0JS40 B4PCU7 A0A182GXQ7 B4J350 A0A1I8M6J3 A0A1L8EEI5 A0A0A9X0D9 A0A0L0CJD1 A0A0K8TPE1 B4KZY6 A0A1Y1NHT9 A0A2R5L9V6 A0A034VFE1 A0A0K8UZB4 A0A1I8PLI3 A0A1A9XBG8 A0A1B0BHC0 A0A224XVH4 D3TLM6 A0A069DR80 A0A0P4VHG1 R4G3D8 W8BXZ2 A0A1B0FB97 A0A162T238 A0A0P5BFZ7 A0A0P6H3S9 A0A1S3D7M0 A0A3L8E095 A0A0N8ARF7 A0A023FP24 A0A0P5E7E4
I4DQE8 A0A194QXW1 A0A2P8Z6P2 A0A1L8E0I2 U5ETX7 A0A2J7Q2L7 A0A1L8E0N6 A0A1B6JRC9 A0A1B0CFY2 A0A182WC31 A0A1B6ERE2 V5IA77 A0A336LKT7 A0A182T7K3 A0A2M4BXB1 A0A182KBE8 A0A182Q115 T1DER2 T1E7A3 A0A182V682 A0A182U1T4 A0A182X904 A0A182LKX4 Q7Q5G0 A0A182I442 A0A1J1IJC4 A0A1B6CKI0 W5JSE8 A0A2M4BX44 A0A182FF92 A0A2M3Z4C6 A0A182IUK7 A0A2M4AAH3 A0A084VAX3 A0A182LV84 A0A182H793 A0A182PB87 A0A182YNV4 D6WIB0 A0A023ENH9 A0A182NJ35 A0A023EME8 A0A067QGG8 A0A182RMP5 Q29E23 B4H416 B0X266 J3JV29 B4HTK8 B4QP26 A0A3B0JZI3 N6TWH4 B4N6C3 A0A1W4W036 A0A0M4EBY7 A0A1A9WLU0 Q9VZU4 A0A1D2N063 B3M7D6 T1PFE4 B4LFE3 B3NF35 A0A3B0JS40 B4PCU7 A0A182GXQ7 B4J350 A0A1I8M6J3 A0A1L8EEI5 A0A0A9X0D9 A0A0L0CJD1 A0A0K8TPE1 B4KZY6 A0A1Y1NHT9 A0A2R5L9V6 A0A034VFE1 A0A0K8UZB4 A0A1I8PLI3 A0A1A9XBG8 A0A1B0BHC0 A0A224XVH4 D3TLM6 A0A069DR80 A0A0P4VHG1 R4G3D8 W8BXZ2 A0A1B0FB97 A0A162T238 A0A0P5BFZ7 A0A0P6H3S9 A0A1S3D7M0 A0A3L8E095 A0A0N8ARF7 A0A023FP24 A0A0P5E7E4
Pubmed
19121390
22118469
26227816
28756777
22651552
26354079
+ More
29403074 24330624 20966253 12364791 14747013 17210077 20920257 23761445 24438588 26483478 25244985 18362917 19820115 24945155 24845553 15632085 17994087 22516182 23537049 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27289101 17550304 25315136 25401762 26823975 26108605 26369729 28004739 25348373 20353571 26334808 27129103 24495485 30249741
29403074 24330624 20966253 12364791 14747013 17210077 20920257 23761445 24438588 26483478 25244985 18362917 19820115 24945155 24845553 15632085 17994087 22516182 23537049 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 27289101 17550304 25315136 25401762 26823975 26108605 26369729 28004739 25348373 20353571 26334808 27129103 24495485 30249741
EMBL
BABH01042366
AGBW02014361
OWR41797.1
NWSH01001174
PCG72252.1
JTDY01000948
+ More
KOB75324.1 KZ149915 PZC77971.1 ODYU01008130 SOQ51477.1 AK404078 KQ459598 BAM20138.1 KPI94494.1 KQ460949 KPJ10378.1 PYGN01000172 PSN52161.1 GFDF01001844 JAV12240.1 GANO01002579 JAB57292.1 NEVH01019369 PNF22819.1 GFDF01001845 JAV12239.1 GECU01033129 GECU01005936 JAS74577.1 JAT01771.1 AJWK01010557 GECZ01029214 JAS40555.1 GALX01001665 JAB66801.1 UFQT01000040 UFQT01001876 SSX18580.1 SSX32067.1 GGFJ01008430 MBW57571.1 AXCN02000458 GALA01000937 JAA93915.1 GAMD01003217 JAA98373.1 AAAB01008960 EAA11886.2 APCN01002272 CVRI01000051 CRK99644.1 GEDC01023376 JAS13922.1 ADMH02000301 ETN67046.1 GGFJ01008431 MBW57572.1 GGFM01002547 MBW23298.1 GGFK01004409 MBW37730.1 ATLV01004724 KE524237 KFB35117.1 AXCM01005863 JXUM01027726 KQ560799 KXJ80847.1 KQ971321 EFA01282.1 GAPW01003112 JAC10486.1 GAPW01003111 JAC10487.1 KK853475 KDR07346.1 CH379070 EAL30240.1 CH479208 EDW31131.1 DS232281 EDS38996.1 BT127095 KB631941 AEE62057.1 ERL87389.1 CH480817 EDW50279.1 CM000363 CM002912 EDX09021.1 KMY97270.1 OUUW01000002 SPP76518.1 APGK01049732 KB741156 ENN73605.1 CH964154 EDW79912.1 CP012525 ALC45066.1 AE014296 AY118532 BT011342 AAF47723.1 AAM49901.1 AAR96134.1 LJIJ01000335 ODM98687.1 CH902618 EDV39834.1 KA647409 AFP62038.1 CH940647 EDW69241.1 CH954178 EDV50377.1 SPP76519.1 CM000159 EDW93851.1 JXUM01095989 KQ564236 KXJ72538.1 CH916366 EDV96121.1 GFDG01001689 JAV17110.1 GBHO01033044 GBRD01016462 GDHC01007661 JAG10560.1 JAG49364.1 JAQ10968.1 JRES01000318 KNC32356.1 GDAI01001592 JAI16011.1 CH933809 EDW17998.1 GEZM01001945 JAV97444.1 GGLE01002156 MBY06282.1 GAKP01018130 JAC40822.1 GDHF01020639 JAI31675.1 JXJN01014257 GFTR01004313 JAW12113.1 EZ422328 ADD18604.1 GBGD01002524 JAC86365.1 GDKW01002696 JAI53899.1 ACPB03020531 GAHY01002290 JAA75220.1 GAMC01000285 JAC06271.1 CCAG010014904 LRGB01000018 KZS21833.1 GDIP01184902 JAJ38500.1 GDIQ01037448 JAN57289.1 QOIP01000002 RLU26134.1 GDIQ01250203 JAK01522.1 GBBK01001000 JAC23482.1 GDIP01145711 JAJ77691.1
KOB75324.1 KZ149915 PZC77971.1 ODYU01008130 SOQ51477.1 AK404078 KQ459598 BAM20138.1 KPI94494.1 KQ460949 KPJ10378.1 PYGN01000172 PSN52161.1 GFDF01001844 JAV12240.1 GANO01002579 JAB57292.1 NEVH01019369 PNF22819.1 GFDF01001845 JAV12239.1 GECU01033129 GECU01005936 JAS74577.1 JAT01771.1 AJWK01010557 GECZ01029214 JAS40555.1 GALX01001665 JAB66801.1 UFQT01000040 UFQT01001876 SSX18580.1 SSX32067.1 GGFJ01008430 MBW57571.1 AXCN02000458 GALA01000937 JAA93915.1 GAMD01003217 JAA98373.1 AAAB01008960 EAA11886.2 APCN01002272 CVRI01000051 CRK99644.1 GEDC01023376 JAS13922.1 ADMH02000301 ETN67046.1 GGFJ01008431 MBW57572.1 GGFM01002547 MBW23298.1 GGFK01004409 MBW37730.1 ATLV01004724 KE524237 KFB35117.1 AXCM01005863 JXUM01027726 KQ560799 KXJ80847.1 KQ971321 EFA01282.1 GAPW01003112 JAC10486.1 GAPW01003111 JAC10487.1 KK853475 KDR07346.1 CH379070 EAL30240.1 CH479208 EDW31131.1 DS232281 EDS38996.1 BT127095 KB631941 AEE62057.1 ERL87389.1 CH480817 EDW50279.1 CM000363 CM002912 EDX09021.1 KMY97270.1 OUUW01000002 SPP76518.1 APGK01049732 KB741156 ENN73605.1 CH964154 EDW79912.1 CP012525 ALC45066.1 AE014296 AY118532 BT011342 AAF47723.1 AAM49901.1 AAR96134.1 LJIJ01000335 ODM98687.1 CH902618 EDV39834.1 KA647409 AFP62038.1 CH940647 EDW69241.1 CH954178 EDV50377.1 SPP76519.1 CM000159 EDW93851.1 JXUM01095989 KQ564236 KXJ72538.1 CH916366 EDV96121.1 GFDG01001689 JAV17110.1 GBHO01033044 GBRD01016462 GDHC01007661 JAG10560.1 JAG49364.1 JAQ10968.1 JRES01000318 KNC32356.1 GDAI01001592 JAI16011.1 CH933809 EDW17998.1 GEZM01001945 JAV97444.1 GGLE01002156 MBY06282.1 GAKP01018130 JAC40822.1 GDHF01020639 JAI31675.1 JXJN01014257 GFTR01004313 JAW12113.1 EZ422328 ADD18604.1 GBGD01002524 JAC86365.1 GDKW01002696 JAI53899.1 ACPB03020531 GAHY01002290 JAA75220.1 GAMC01000285 JAC06271.1 CCAG010014904 LRGB01000018 KZS21833.1 GDIP01184902 JAJ38500.1 GDIQ01037448 JAN57289.1 QOIP01000002 RLU26134.1 GDIQ01250203 JAK01522.1 GBBK01001000 JAC23482.1 GDIP01145711 JAJ77691.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000037510
UP000053268
UP000053240
+ More
UP000245037 UP000235965 UP000092461 UP000075920 UP000075901 UP000075881 UP000075886 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000183832 UP000000673 UP000069272 UP000075880 UP000030765 UP000075883 UP000069940 UP000249989 UP000075885 UP000076408 UP000007266 UP000075884 UP000027135 UP000075900 UP000001819 UP000008744 UP000002320 UP000030742 UP000001292 UP000000304 UP000268350 UP000019118 UP000007798 UP000192221 UP000092553 UP000091820 UP000000803 UP000094527 UP000007801 UP000008792 UP000008711 UP000002282 UP000001070 UP000095301 UP000037069 UP000009192 UP000095300 UP000092443 UP000092460 UP000015103 UP000092444 UP000076858 UP000079169 UP000279307
UP000245037 UP000235965 UP000092461 UP000075920 UP000075901 UP000075881 UP000075886 UP000075903 UP000075902 UP000076407 UP000075882 UP000007062 UP000075840 UP000183832 UP000000673 UP000069272 UP000075880 UP000030765 UP000075883 UP000069940 UP000249989 UP000075885 UP000076408 UP000007266 UP000075884 UP000027135 UP000075900 UP000001819 UP000008744 UP000002320 UP000030742 UP000001292 UP000000304 UP000268350 UP000019118 UP000007798 UP000192221 UP000092553 UP000091820 UP000000803 UP000094527 UP000007801 UP000008792 UP000008711 UP000002282 UP000001070 UP000095301 UP000037069 UP000009192 UP000095300 UP000092443 UP000092460 UP000015103 UP000092444 UP000076858 UP000079169 UP000279307
PRIDE
Pfam
PF00329 Complex1_30kDa
Interpro
SUPFAM
SSF143243
SSF143243
Gene 3D
ProteinModelPortal
H9JY65
A0A212EJY1
A0A2A4JLG1
A0A0L7LIY8
A0A2W1BUD9
A0A2H1WEN3
+ More
I4DQE8 A0A194QXW1 A0A2P8Z6P2 A0A1L8E0I2 U5ETX7 A0A2J7Q2L7 A0A1L8E0N6 A0A1B6JRC9 A0A1B0CFY2 A0A182WC31 A0A1B6ERE2 V5IA77 A0A336LKT7 A0A182T7K3 A0A2M4BXB1 A0A182KBE8 A0A182Q115 T1DER2 T1E7A3 A0A182V682 A0A182U1T4 A0A182X904 A0A182LKX4 Q7Q5G0 A0A182I442 A0A1J1IJC4 A0A1B6CKI0 W5JSE8 A0A2M4BX44 A0A182FF92 A0A2M3Z4C6 A0A182IUK7 A0A2M4AAH3 A0A084VAX3 A0A182LV84 A0A182H793 A0A182PB87 A0A182YNV4 D6WIB0 A0A023ENH9 A0A182NJ35 A0A023EME8 A0A067QGG8 A0A182RMP5 Q29E23 B4H416 B0X266 J3JV29 B4HTK8 B4QP26 A0A3B0JZI3 N6TWH4 B4N6C3 A0A1W4W036 A0A0M4EBY7 A0A1A9WLU0 Q9VZU4 A0A1D2N063 B3M7D6 T1PFE4 B4LFE3 B3NF35 A0A3B0JS40 B4PCU7 A0A182GXQ7 B4J350 A0A1I8M6J3 A0A1L8EEI5 A0A0A9X0D9 A0A0L0CJD1 A0A0K8TPE1 B4KZY6 A0A1Y1NHT9 A0A2R5L9V6 A0A034VFE1 A0A0K8UZB4 A0A1I8PLI3 A0A1A9XBG8 A0A1B0BHC0 A0A224XVH4 D3TLM6 A0A069DR80 A0A0P4VHG1 R4G3D8 W8BXZ2 A0A1B0FB97 A0A162T238 A0A0P5BFZ7 A0A0P6H3S9 A0A1S3D7M0 A0A3L8E095 A0A0N8ARF7 A0A023FP24 A0A0P5E7E4
I4DQE8 A0A194QXW1 A0A2P8Z6P2 A0A1L8E0I2 U5ETX7 A0A2J7Q2L7 A0A1L8E0N6 A0A1B6JRC9 A0A1B0CFY2 A0A182WC31 A0A1B6ERE2 V5IA77 A0A336LKT7 A0A182T7K3 A0A2M4BXB1 A0A182KBE8 A0A182Q115 T1DER2 T1E7A3 A0A182V682 A0A182U1T4 A0A182X904 A0A182LKX4 Q7Q5G0 A0A182I442 A0A1J1IJC4 A0A1B6CKI0 W5JSE8 A0A2M4BX44 A0A182FF92 A0A2M3Z4C6 A0A182IUK7 A0A2M4AAH3 A0A084VAX3 A0A182LV84 A0A182H793 A0A182PB87 A0A182YNV4 D6WIB0 A0A023ENH9 A0A182NJ35 A0A023EME8 A0A067QGG8 A0A182RMP5 Q29E23 B4H416 B0X266 J3JV29 B4HTK8 B4QP26 A0A3B0JZI3 N6TWH4 B4N6C3 A0A1W4W036 A0A0M4EBY7 A0A1A9WLU0 Q9VZU4 A0A1D2N063 B3M7D6 T1PFE4 B4LFE3 B3NF35 A0A3B0JS40 B4PCU7 A0A182GXQ7 B4J350 A0A1I8M6J3 A0A1L8EEI5 A0A0A9X0D9 A0A0L0CJD1 A0A0K8TPE1 B4KZY6 A0A1Y1NHT9 A0A2R5L9V6 A0A034VFE1 A0A0K8UZB4 A0A1I8PLI3 A0A1A9XBG8 A0A1B0BHC0 A0A224XVH4 D3TLM6 A0A069DR80 A0A0P4VHG1 R4G3D8 W8BXZ2 A0A1B0FB97 A0A162T238 A0A0P5BFZ7 A0A0P6H3S9 A0A1S3D7M0 A0A3L8E095 A0A0N8ARF7 A0A023FP24 A0A0P5E7E4
PDB
5XTI
E-value=1.68636e-86,
Score=812
Ontologies
PATHWAY
GO
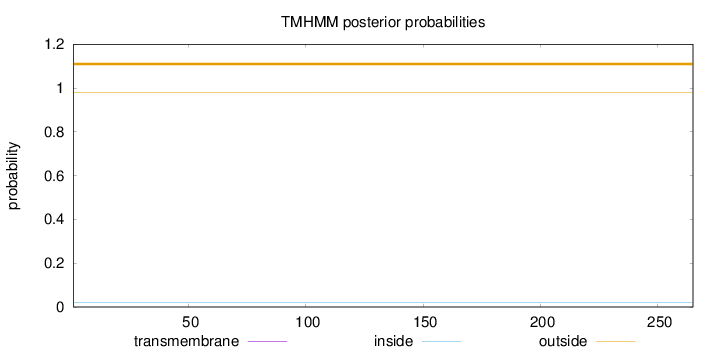
Topology
Length:
265
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000470000000000001
Exp number, first 60 AAs:
0.00024
Total prob of N-in:
0.01994
outside
1 - 265
Population Genetic Test Statistics
Pi
7.482212
Theta
6.521812
Tajima's D
0.489196
CLR
0.30762
CSRT
0.530773461326934
Interpretation
Uncertain
