Pre Gene Modal
BGIBMGA000189
Annotation
PREDICTED:_protein_VAC14_homolog_isoform_X2_[Bombyx_mori]
Full name
Protein VAC14 homolog
Location in the cell
Cytoplasmic Reliability : 2.099
Sequence
CDS
ATGTCCGAGCGAGACTACGCACCGCTGAGCATAGCTTGTGTTCGTGGGCTTTGCGACAAATTATATGAGAAGAGAAAAGTTGCTGGAGTCGAGATTGAGAAGATGGTGAAAGATTTTAATGATGCAAAAAATACTAGCCAAATAAAAAAATTGATCAAAGTCTTGGGCCAGGACTTGATGTCTTCAACTAATCCTAATATGAAAAACGGAGCACTTATGGGTCTGTCTACTGTAGCAGTGGGGCTTGGCAAGGCATCAGTAGATTACTTACCAGAATTAACAAATCCAATAGTGGCTTGCTTCAGCGAGAGCGAATCTCGTGTAAGGTACCAAGCAGCTGAGGCATTGTTCAACGTGCTGAAGATTGTCCGAGGCGCTTCTTTATCACAGTTCCCTGTAATTTTCGATGCGCTGGCTAAACTCGCTGCTGATCCTGAGCAGCAGGTGAAGCATGCTGCTGAATTGTTGGATAGATTGCTCAAGGACATCGTGACCGAGAGCGGGTCCGTGGAGCTGTCGTCGCTGGTGGGGCTGGTGCGAGAGCGCATGTACACGCGCTCGGCGCCGGCGCGCCAGCTCGCCGTGTCCTGGGTGGCGGTGCTGGACGCCGTGCCCGACCTCAACGTGCTGGCCCATCTCCCGGACTTACTGGACGGGCTCTTCAAGATGCTAGACGATCCTAACCCGGAGATCAGAAGGATGTGCGATGTCCAGCTGAACGAGTTCCTCCGCAGCATTAAGAAAGAGCCAGCCAAAGTAGACTTCCAAGCCATGATCAACATCCTTATCATACACGCGCAGTCGCCCGAGGAACTGCTGCAGCTGACGGCGATTACGTGGCTCAAGGAGTTCGTGGAGCTGGCCGGCGCCGCCATGCTGCCCTTCGCCTCGGGCATCCTGTGCGCCGTGCTGCCCTGCCACGCATACTCCGACGAGCCAAGGAAGAGCATACGGGAGACGGCGGCCACGGTCAACTTTCAACTGATCAAACTGTTGGCTGCCGAGAGCGAGGAGGCTGGTGGTGAGGAGGCGGCGGGCGAGGAGGGGGGCGCGCTCAACCTGGAGGCCGTGGTGGGGGTGCTGACGCAGATGCTGCACCACAGCTCCGTGCACACTAAAGTGGCCGCCCTGGATTGGATCCTTCACTTATACAATAAGCTCCCCAAACAGATGTGCGGCGAGACGGAGCGCGTGTGGGCGGGCGCGGTGGGCAGCCTCACGGACGCGGCGCACGACGTGCTGCGGCGCGCGCTCGCCGTGCTGGCAGAGATCTGCTCCGCCGCCGGAGATTTGCAGTCTAGTCCTTACTACTACAAGTTCCTACAAGCTTTATTGAGGCTTTTGGCGGCAGACGAGAACCTGTTGGAAGACAGAGGAGCTTTCATTATCAGGCAGCTGTGCGTCCTGCTGAACGCGGAGGACATATACAGGGCCCTAGCGAACATTCTGCAGCAGGAGAAGAATCTCCGCTTCGTGACCACCATGGTGGACATCCTCAACACCATACTGCTGACGTCCGCCGAGCTATACGAGCTGCGGGCGCTGCTGAAGGACCGCAGCAAGCCGTGGAGCCGCTCGCTGTTCCTGGCGCTGCACGGCTGCTGGTGCCACAGCCCCGTGTCGCTGCTGGCGCTGTGTCTGCTGGCGCACCACTACCGGCACTGCAACACGCTCATCGCCACCTTCGGCGACCTCGAGATAACCGTGGAGTTCCTGACGGAGGTGGACAAGCTGGTGCAGCTCATTGAGTCGCCCATATTCGCCTACCTGCGCCTGGAGCTGGTGGGCGAGCACAGCGCCGGCCTGCGCGCCGCGCTCCGCACCGCCCGCGGTACGGACAACCAACTACCTTCCTAA
Protein
MSERDYAPLSIACVRGLCDKLYEKRKVAGVEIEKMVKDFNDAKNTSQIKKLIKVLGQDLMSSTNPNMKNGALMGLSTVAVGLGKASVDYLPELTNPIVACFSESESRVRYQAAEALFNVLKIVRGASLSQFPVIFDALAKLAADPEQQVKHAAELLDRLLKDIVTESGSVELSSLVGLVRERMYTRSAPARQLAVSWVAVLDAVPDLNVLAHLPDLLDGLFKMLDDPNPEIRRMCDVQLNEFLRSIKKEPAKVDFQAMINILIIHAQSPEELLQLTAITWLKEFVELAGAAMLPFASGILCAVLPCHAYSDEPRKSIRETAATVNFQLIKLLAAESEEAGGEEAAGEEGGALNLEAVVGVLTQMLHHSSVHTKVAALDWILHLYNKLPKQMCGETERVWAGAVGSLTDAAHDVLRRALAVLAEICSAAGDLQSSPYYYKFLQALLRLLAADENLLEDRGAFIIRQLCVLLNAEDIYRALANILQQEKNLRFVTTMVDILNTILLTSAELYELRALLKDRSKPWSRSLFLALHGCWCHSPVSLLALCLLAHHYRHCNTLIATFGDLEITVEFLTEVDKLVQLIESPIFAYLRLELVGEHSAGLRAALRTARGTDNQLPS
Summary
Description
The PI(3,5)P2 regulatory complex regulates both the synthesis and turnover of phosphatidylinositol 3,5-bisphosphate (PtdIns(3,5)P2). Acts as a positive activator of PIKfyve kinase activity (By similarity).
Subunit
Component of the PI(3,5)P2 regulatory complex/PAS complex, at least composed of PIKFYVE, FIG4 and VAC14. VAC14 nucleates the assembly of the complex and serves as a scaffold (By similarity).
Similarity
Belongs to the VAC14 family.
Keywords
Complete proteome
Endosome
Membrane
Reference proteome
Repeat
Feature
chain Protein VAC14 homolog
Uniprot
H9ISG2
A0A212EK04
D6WPN6
A0A154PKW1
E2BXH5
A0A1Y1MAW0
+ More
A0A139WEA7 A0A151WJQ9 A0A0L7QNU0 A0A067QRT8 E2AC17 A0A2A3ELH5 F4X110 A0A195CKL6 A0A158NI19 A0A0J7L0B3 E0VQV6 A0A2J7R3P0 A0A1B6I0P4 A0A1Y1M3R9 A0A1B6LBW9 A0A026WYQ0 A0A0M8ZSI6 A0A2J7R3M5 A0A1W4X449 A0A151I4L1 A0A232FKC3 K7IZD8 A0A1D2N298 A0A195F638 A0A0P6B9K8 A0A0P5HIL8 A0A0P5HRA7 A0A1B6E9H2 A0A164Z729 A0A0P6IEE6 A0A147BD08 N6T3G4 A0A195EFY1 A0A2R5LB63 A0A0L7L1D0 A0A293MXF6 A0A023F269 U4UQF3 A0A069DWW3 A0A0A9X6L4 A0A1B0CQ03 A0A131YIN1 V4BVL4 A0A224ZBK8 A0A087TV22 W8CD00 A0A2L2YQZ5 A0A0P5D0A6 F1NWG6 A0A2T7NI13 A0A226P2M0 Q5ZIW5 F6U0G4 A0A1I8MR22 A0A0P5CLR2 A0A0A1XM24 T2MEF9 B4MBI2 A0A151MSN0 A0A0P5DFF2 A0A226MPI1 A0A182N0G7 G1S284 A0A2H8TUV5 A0A182Q8M2 J9JVA2 H2NRF6 A0A2K6KTU5 A0A0L0CBX0 E9GAP5 A0A0D9QWF5 A0A2K5ESF7 A0A1I8PF19 A0A2K6KTT9 A0A2K6RIM8 A0A2K6RIM0 A0A0K8U4B4 A0A034W6B7 A0A182P6U2 B3P4A7 A0A2K5R1D8 A0A0M4F3J9 Q16Q69 A0A084VV66 A0A182W468 A0A336M062 A0A182I9J8 Q7QE90 A0A182JYM5 A0A182V5G1 B4QS97 A0A182MFN2
A0A139WEA7 A0A151WJQ9 A0A0L7QNU0 A0A067QRT8 E2AC17 A0A2A3ELH5 F4X110 A0A195CKL6 A0A158NI19 A0A0J7L0B3 E0VQV6 A0A2J7R3P0 A0A1B6I0P4 A0A1Y1M3R9 A0A1B6LBW9 A0A026WYQ0 A0A0M8ZSI6 A0A2J7R3M5 A0A1W4X449 A0A151I4L1 A0A232FKC3 K7IZD8 A0A1D2N298 A0A195F638 A0A0P6B9K8 A0A0P5HIL8 A0A0P5HRA7 A0A1B6E9H2 A0A164Z729 A0A0P6IEE6 A0A147BD08 N6T3G4 A0A195EFY1 A0A2R5LB63 A0A0L7L1D0 A0A293MXF6 A0A023F269 U4UQF3 A0A069DWW3 A0A0A9X6L4 A0A1B0CQ03 A0A131YIN1 V4BVL4 A0A224ZBK8 A0A087TV22 W8CD00 A0A2L2YQZ5 A0A0P5D0A6 F1NWG6 A0A2T7NI13 A0A226P2M0 Q5ZIW5 F6U0G4 A0A1I8MR22 A0A0P5CLR2 A0A0A1XM24 T2MEF9 B4MBI2 A0A151MSN0 A0A0P5DFF2 A0A226MPI1 A0A182N0G7 G1S284 A0A2H8TUV5 A0A182Q8M2 J9JVA2 H2NRF6 A0A2K6KTU5 A0A0L0CBX0 E9GAP5 A0A0D9QWF5 A0A2K5ESF7 A0A1I8PF19 A0A2K6KTT9 A0A2K6RIM8 A0A2K6RIM0 A0A0K8U4B4 A0A034W6B7 A0A182P6U2 B3P4A7 A0A2K5R1D8 A0A0M4F3J9 Q16Q69 A0A084VV66 A0A182W468 A0A336M062 A0A182I9J8 Q7QE90 A0A182JYM5 A0A182V5G1 B4QS97 A0A182MFN2
Pubmed
19121390
22118469
18362917
19820115
20798317
28004739
+ More
24845553 21719571 21347285 20566863 24508170 30249741 28648823 20075255 27289101 29652888 23537049 26227816 25474469 26334808 25401762 26823975 26830274 23254933 28797301 24495485 26561354 15592404 24621616 15642098 17431167 25315136 25830018 24065732 17994087 22293439 26108605 21292972 25362486 25348373 17510324 24438588 12364791 14747013 17210077
24845553 21719571 21347285 20566863 24508170 30249741 28648823 20075255 27289101 29652888 23537049 26227816 25474469 26334808 25401762 26823975 26830274 23254933 28797301 24495485 26561354 15592404 24621616 15642098 17431167 25315136 25830018 24065732 17994087 22293439 26108605 21292972 25362486 25348373 17510324 24438588 12364791 14747013 17210077
EMBL
BABH01040702
BABH01040703
AGBW02014361
OWR41800.1
KQ971354
EFA06197.2
+ More
KQ434938 KZC12114.1 GL451254 EFN79623.1 GEZM01041344 JAV80497.1 KYB26276.1 KQ983039 KYQ48064.1 KQ414855 KOC60156.1 KK853035 KDR12240.1 GL438389 EFN69035.1 KZ288223 PBC32009.1 GL888520 EGI59870.1 KQ977622 KYN01270.1 ADTU01016184 ADTU01016185 LBMM01001650 KMQ95944.1 DS235442 EEB15762.1 NEVH01007819 PNF35435.1 GECU01027261 JAS80445.1 GEZM01041346 JAV80494.1 GEBQ01018798 JAT21179.1 KK107078 QOIP01000011 EZA60269.1 RLU16468.1 KQ435867 KOX70260.1 PNF35433.1 KQ976456 KYM84873.1 NNAY01000078 OXU31191.1 LJIJ01000285 ODM99378.1 KQ981763 KYN36055.1 GDIP01024456 JAM79259.1 GDIQ01226943 JAK24782.1 GDIQ01223442 JAK28283.1 GEDC01002728 JAS34570.1 LRGB01000781 KZS16018.1 GDIQ01012602 JAN82135.1 GEGO01006758 JAR88646.1 APGK01045645 KB741037 ENN74689.1 KQ978957 KYN27148.1 GGLE01002582 MBY06708.1 JTDY01003648 KOB69220.1 GFWV01020071 MAA44799.1 GBBI01003030 JAC15682.1 KB632319 ERL92270.1 GBGD01000692 JAC88197.1 GBHO01030884 GBRD01016792 GDHC01008895 GDHC01008399 JAG12720.1 JAG49035.1 JAQ09734.1 JAQ10230.1 AJWK01022899 GEDV01010162 JAP78395.1 KB201977 ESO93069.1 GFPF01013268 MAA24414.1 KK116858 KFM68961.1 GAMC01002811 JAC03745.1 IAAA01041532 LAA10503.1 GDIP01164638 JAJ58764.1 AADN05000561 PZQS01000012 PVD20795.1 AWGT02000192 OXB74071.1 AJ720669 JSUE03026554 JSUE03026555 GDIP01168675 JAJ54727.1 GBXI01002382 JAD11910.1 HAAD01004078 CDG70310.1 CH940656 EDW58453.1 AKHW03005169 KYO27514.1 GDIP01158293 JAJ65109.1 MCFN01000563 OXB57223.1 ADFV01127321 ADFV01127322 ADFV01127323 ADFV01127324 ADFV01127325 ADFV01127326 ADFV01127327 GFXV01005925 MBW17730.1 AXCN02001412 ABLF02035835 ABGA01059396 ABGA01059397 ABGA01059398 ABGA01059399 ABGA01059400 ABGA01059401 ABGA01059402 ABGA01059403 ABGA01059404 ABGA01059405 NDHI03003408 PNJ61738.1 JRES01000634 KNC29750.1 GL732537 EFX83292.1 AQIB01129532 GDHF01031169 JAI21145.1 GAKP01009639 JAC49313.1 CH954181 EDV49352.1 CP012526 ALC45820.1 CH477759 EAT36537.1 ATLV01017157 KE525157 KFB41860.1 UFQT01000350 SSX23450.1 APCN01000987 AAAB01008847 EAA06890.5 CM000364 EDX13193.1 AXCM01001219
KQ434938 KZC12114.1 GL451254 EFN79623.1 GEZM01041344 JAV80497.1 KYB26276.1 KQ983039 KYQ48064.1 KQ414855 KOC60156.1 KK853035 KDR12240.1 GL438389 EFN69035.1 KZ288223 PBC32009.1 GL888520 EGI59870.1 KQ977622 KYN01270.1 ADTU01016184 ADTU01016185 LBMM01001650 KMQ95944.1 DS235442 EEB15762.1 NEVH01007819 PNF35435.1 GECU01027261 JAS80445.1 GEZM01041346 JAV80494.1 GEBQ01018798 JAT21179.1 KK107078 QOIP01000011 EZA60269.1 RLU16468.1 KQ435867 KOX70260.1 PNF35433.1 KQ976456 KYM84873.1 NNAY01000078 OXU31191.1 LJIJ01000285 ODM99378.1 KQ981763 KYN36055.1 GDIP01024456 JAM79259.1 GDIQ01226943 JAK24782.1 GDIQ01223442 JAK28283.1 GEDC01002728 JAS34570.1 LRGB01000781 KZS16018.1 GDIQ01012602 JAN82135.1 GEGO01006758 JAR88646.1 APGK01045645 KB741037 ENN74689.1 KQ978957 KYN27148.1 GGLE01002582 MBY06708.1 JTDY01003648 KOB69220.1 GFWV01020071 MAA44799.1 GBBI01003030 JAC15682.1 KB632319 ERL92270.1 GBGD01000692 JAC88197.1 GBHO01030884 GBRD01016792 GDHC01008895 GDHC01008399 JAG12720.1 JAG49035.1 JAQ09734.1 JAQ10230.1 AJWK01022899 GEDV01010162 JAP78395.1 KB201977 ESO93069.1 GFPF01013268 MAA24414.1 KK116858 KFM68961.1 GAMC01002811 JAC03745.1 IAAA01041532 LAA10503.1 GDIP01164638 JAJ58764.1 AADN05000561 PZQS01000012 PVD20795.1 AWGT02000192 OXB74071.1 AJ720669 JSUE03026554 JSUE03026555 GDIP01168675 JAJ54727.1 GBXI01002382 JAD11910.1 HAAD01004078 CDG70310.1 CH940656 EDW58453.1 AKHW03005169 KYO27514.1 GDIP01158293 JAJ65109.1 MCFN01000563 OXB57223.1 ADFV01127321 ADFV01127322 ADFV01127323 ADFV01127324 ADFV01127325 ADFV01127326 ADFV01127327 GFXV01005925 MBW17730.1 AXCN02001412 ABLF02035835 ABGA01059396 ABGA01059397 ABGA01059398 ABGA01059399 ABGA01059400 ABGA01059401 ABGA01059402 ABGA01059403 ABGA01059404 ABGA01059405 NDHI03003408 PNJ61738.1 JRES01000634 KNC29750.1 GL732537 EFX83292.1 AQIB01129532 GDHF01031169 JAI21145.1 GAKP01009639 JAC49313.1 CH954181 EDV49352.1 CP012526 ALC45820.1 CH477759 EAT36537.1 ATLV01017157 KE525157 KFB41860.1 UFQT01000350 SSX23450.1 APCN01000987 AAAB01008847 EAA06890.5 CM000364 EDX13193.1 AXCM01001219
Proteomes
UP000005204
UP000007151
UP000007266
UP000076502
UP000008237
UP000075809
+ More
UP000053825 UP000027135 UP000000311 UP000242457 UP000007755 UP000078542 UP000005205 UP000036403 UP000009046 UP000235965 UP000053097 UP000279307 UP000053105 UP000192223 UP000078540 UP000215335 UP000002358 UP000094527 UP000078541 UP000076858 UP000019118 UP000078492 UP000037510 UP000030742 UP000092461 UP000030746 UP000054359 UP000000539 UP000245119 UP000198419 UP000006718 UP000095301 UP000008792 UP000050525 UP000198323 UP000075884 UP000001073 UP000075886 UP000007819 UP000001595 UP000233180 UP000037069 UP000000305 UP000029965 UP000233020 UP000095300 UP000233200 UP000075885 UP000008711 UP000233040 UP000092553 UP000008820 UP000030765 UP000075920 UP000075840 UP000007062 UP000075881 UP000075903 UP000000304 UP000075883
UP000053825 UP000027135 UP000000311 UP000242457 UP000007755 UP000078542 UP000005205 UP000036403 UP000009046 UP000235965 UP000053097 UP000279307 UP000053105 UP000192223 UP000078540 UP000215335 UP000002358 UP000094527 UP000078541 UP000076858 UP000019118 UP000078492 UP000037510 UP000030742 UP000092461 UP000030746 UP000054359 UP000000539 UP000245119 UP000198419 UP000006718 UP000095301 UP000008792 UP000050525 UP000198323 UP000075884 UP000001073 UP000075886 UP000007819 UP000001595 UP000233180 UP000037069 UP000000305 UP000029965 UP000233020 UP000095300 UP000233200 UP000075885 UP000008711 UP000233040 UP000092553 UP000008820 UP000030765 UP000075920 UP000075840 UP000007062 UP000075881 UP000075903 UP000000304 UP000075883
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
H9ISG2
A0A212EK04
D6WPN6
A0A154PKW1
E2BXH5
A0A1Y1MAW0
+ More
A0A139WEA7 A0A151WJQ9 A0A0L7QNU0 A0A067QRT8 E2AC17 A0A2A3ELH5 F4X110 A0A195CKL6 A0A158NI19 A0A0J7L0B3 E0VQV6 A0A2J7R3P0 A0A1B6I0P4 A0A1Y1M3R9 A0A1B6LBW9 A0A026WYQ0 A0A0M8ZSI6 A0A2J7R3M5 A0A1W4X449 A0A151I4L1 A0A232FKC3 K7IZD8 A0A1D2N298 A0A195F638 A0A0P6B9K8 A0A0P5HIL8 A0A0P5HRA7 A0A1B6E9H2 A0A164Z729 A0A0P6IEE6 A0A147BD08 N6T3G4 A0A195EFY1 A0A2R5LB63 A0A0L7L1D0 A0A293MXF6 A0A023F269 U4UQF3 A0A069DWW3 A0A0A9X6L4 A0A1B0CQ03 A0A131YIN1 V4BVL4 A0A224ZBK8 A0A087TV22 W8CD00 A0A2L2YQZ5 A0A0P5D0A6 F1NWG6 A0A2T7NI13 A0A226P2M0 Q5ZIW5 F6U0G4 A0A1I8MR22 A0A0P5CLR2 A0A0A1XM24 T2MEF9 B4MBI2 A0A151MSN0 A0A0P5DFF2 A0A226MPI1 A0A182N0G7 G1S284 A0A2H8TUV5 A0A182Q8M2 J9JVA2 H2NRF6 A0A2K6KTU5 A0A0L0CBX0 E9GAP5 A0A0D9QWF5 A0A2K5ESF7 A0A1I8PF19 A0A2K6KTT9 A0A2K6RIM8 A0A2K6RIM0 A0A0K8U4B4 A0A034W6B7 A0A182P6U2 B3P4A7 A0A2K5R1D8 A0A0M4F3J9 Q16Q69 A0A084VV66 A0A182W468 A0A336M062 A0A182I9J8 Q7QE90 A0A182JYM5 A0A182V5G1 B4QS97 A0A182MFN2
A0A139WEA7 A0A151WJQ9 A0A0L7QNU0 A0A067QRT8 E2AC17 A0A2A3ELH5 F4X110 A0A195CKL6 A0A158NI19 A0A0J7L0B3 E0VQV6 A0A2J7R3P0 A0A1B6I0P4 A0A1Y1M3R9 A0A1B6LBW9 A0A026WYQ0 A0A0M8ZSI6 A0A2J7R3M5 A0A1W4X449 A0A151I4L1 A0A232FKC3 K7IZD8 A0A1D2N298 A0A195F638 A0A0P6B9K8 A0A0P5HIL8 A0A0P5HRA7 A0A1B6E9H2 A0A164Z729 A0A0P6IEE6 A0A147BD08 N6T3G4 A0A195EFY1 A0A2R5LB63 A0A0L7L1D0 A0A293MXF6 A0A023F269 U4UQF3 A0A069DWW3 A0A0A9X6L4 A0A1B0CQ03 A0A131YIN1 V4BVL4 A0A224ZBK8 A0A087TV22 W8CD00 A0A2L2YQZ5 A0A0P5D0A6 F1NWG6 A0A2T7NI13 A0A226P2M0 Q5ZIW5 F6U0G4 A0A1I8MR22 A0A0P5CLR2 A0A0A1XM24 T2MEF9 B4MBI2 A0A151MSN0 A0A0P5DFF2 A0A226MPI1 A0A182N0G7 G1S284 A0A2H8TUV5 A0A182Q8M2 J9JVA2 H2NRF6 A0A2K6KTU5 A0A0L0CBX0 E9GAP5 A0A0D9QWF5 A0A2K5ESF7 A0A1I8PF19 A0A2K6KTT9 A0A2K6RIM8 A0A2K6RIM0 A0A0K8U4B4 A0A034W6B7 A0A182P6U2 B3P4A7 A0A2K5R1D8 A0A0M4F3J9 Q16Q69 A0A084VV66 A0A182W468 A0A336M062 A0A182I9J8 Q7QE90 A0A182JYM5 A0A182V5G1 B4QS97 A0A182MFN2
Ontologies
KEGG
GO
PANTHER
Topology
Subcellular location
Endosome membrane
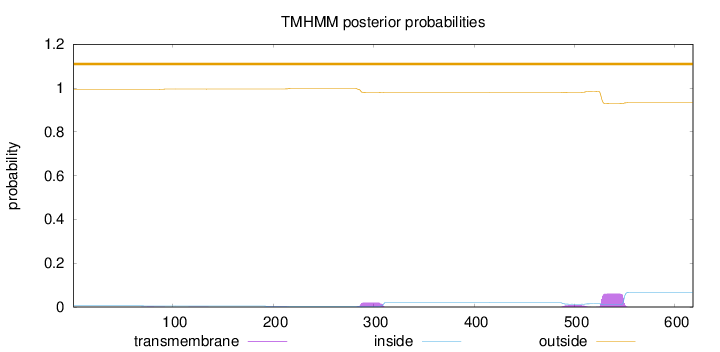
Length:
618
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.11232
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00591
outside
1 - 618
Population Genetic Test Statistics
Pi
7.117504
Theta
12.855282
Tajima's D
-0.595289
CLR
0.095795
CSRT
0.217339133043348
Interpretation
Possibly Positive selection
