Gene
KWMTBOMO11773
Pre Gene Modal
BGIBMGA000200
Annotation
PREDICTED:_E3_ubiquitin-protein_ligase_hrd-like_protein_1_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 3.174
Sequence
CDS
ATGCCCGGGACTTTAGTAGACAGGCTGCCCTTGCCGAATCTCAAGGTTTACACAGCGGGAAGTGTTTTACTTCTTTCAATAGCTGTCTATCATGCCACGATTGTGACTAGTGATCCAAACTGGCGCGCCAATATCACTCTACAGCGTCAGGAAGCATTGGCTGAGCCTGATGGAGAGGCGCAGCTGGTAGACCAAGCTGAAGTAATGCCGGCGCTAAACCTGAATGCCACCAGGAACTTGAACGAGCGCATGGTGGTAATAATAACCTTCATGATGCAGGAACCGCTTTGTATGTGGACATTAATAAACATAGCGTACTGCTTCCTCGCCGTCTTCGGTTTCCTGATCCAGCGCGTCGTCTTCGGTCAGCTCCGGGTCTCTGAGGCTCAGCGCGTCAAGGACAAGTTCTGGAACTATGTCTTCTACAAATTCATCTTCATCTTCGGAGTAATCAACGTTCAGTACATGGACGAGGTGATGCTGTGGTGTTCTTGGTTTACTCTCGTCGGCTTCCTGCACCTGATGGGGCAGCTGTGCAAGGATCGATTTGAATATCTAGCGTCGTCGCCCAACACAGCACGTTGGGTGCATGTTCGACTGGTCGGTCTACTCCTGGGTATACTGGCCGCCTCCACTGCCTTATTGGGCGTTGCAGTCGCATGGGGCTTGCCCGCCGGAAAGGATACTTTCGCGTTTATGGCTGCTGAATGCCTGCTGGTGCTGATGTCGGTGCTGCACGTGGGCGCGCGCTACGTGCTGCAGCAGCGCGGCGCGGACGCGGCCGGCGCGCTCGCCTACTACACGCACCTCGGCTTCGATTCGGCGTCCCTTGTGCTCGAGCTGATGCACGTGGTCCACATGGTGGTGTACAGCAACATGTTGGTGTCGATGGCGTCGCTGGTGCTGCTCATGCAGCTCCGCCACCTGCTGCACGAGCTGCAGGCGCGGCTGCGGAGGCACCGCCTCTACACATCGCTCGCGGACCACATGAGTCGCAATTACCCAATGGCAAGCAAAGAGGAGGTTGAAAAGAATCAAGACAATTGTGCAATCTGCTGGGAACCGATGAAGGAAGCTAGGAAACTGCCCTGTTCTCATCTCTTCCACAACTCGTGCCTATGCCAGTGGGTGCAGCAGGACGCGTCGTGCCCCACGTGCCGCCGCGGCCTCGCACACGGCGCGCCCCCGCCCCCGCCCCCAACCCCGCCGCACAACCACCTCTTCCACTTCGACGGCTCCCGCTACGTGTCGTGGCTGCCCAGCTTCTCCGTGGAGGTGACCCGCGCCCGGCCCTCCGCCGCGCAGCTGGACGCCATGGTGGCGCAGGTGCTGGGCATGTTCCCGCAGGCTGACCCCGCCGCCCTGCGCGCCGACCTGCTGCTCACCCGCTCCGCGCACCTCACGCTCGACAACATCCTCGAGGGCAGGCTGCAGCCGCCCGCAGAAGCACCGACAGTAAGACCGATCTCGCCCCTCGAGGCGCCCCCCGCCCCGGAGCCCGCGCCCCCCGCTCCGGAGCCTGCGCCCCCCGCCGCGGAGCCCGGCCTGGACAATCTAGACGTCGGGTCGACCAGCGCTATATTCTCGAGAAACGCGCCCGAACGAGAACAGAACCTGAGACGCAGGAAGGAGATGCTGGTGGCGGCGGCGCGCAGGCGGTACCTCGAGCGGCGCGGCGAGGCGGCGCCACCACGCAGCTGA
Protein
MPGTLVDRLPLPNLKVYTAGSVLLLSIAVYHATIVTSDPNWRANITLQRQEALAEPDGEAQLVDQAEVMPALNLNATRNLNERMVVIITFMMQEPLCMWTLINIAYCFLAVFGFLIQRVVFGQLRVSEAQRVKDKFWNYVFYKFIFIFGVINVQYMDEVMLWCSWFTLVGFLHLMGQLCKDRFEYLASSPNTARWVHVRLVGLLLGILAASTALLGVAVAWGLPAGKDTFAFMAAECLLVLMSVLHVGARYVLQQRGADAAGALAYYTHLGFDSASLVLELMHVVHMVVYSNMLVSMASLVLLMQLRHLLHELQARLRRHRLYTSLADHMSRNYPMASKEEVEKNQDNCAICWEPMKEARKLPCSHLFHNSCLCQWVQQDASCPTCRRGLAHGAPPPPPPTPPHNHLFHFDGSRYVSWLPSFSVEVTRARPSAAQLDAMVAQVLGMFPQADPAALRADLLLTRSAHLTLDNILEGRLQPPAEAPTVRPISPLEAPPAPEPAPPAPEPAPPAAEPGLDNLDVGSTSAIFSRNAPEREQNLRRRKEMLVAAARRRYLERRGEAAPPRS
Summary
Uniprot
A0A2W1BZ63
A0A194PP26
A0A2J7PI11
A0A1B6GPE6
A0A1B6IRM5
A0A067RD56
+ More
D6WPF0 A0A212EJZ6 A0A1B6E6P4 A0A1Y1L8E1 V9IK44 A0A0M8ZQ75 A0A1Z5LFT8 A0A0T6BAV7 A0A293LCR4 A0A1L8DSF2 A0A1L8DSA8 A0A1Z5L2N8 A0A2R5LBR3 A0A131YXB9 A0A224YUB1 A0A151X5K0 A0A0L7R0D4 A0A2I9LNQ8 A0A147BG56 A0A026WKE0 A0A023FD51 A0A195DRL1 E2BPI8 A0A195EVS6 F4X271 A0A158NAK6 A0A0C9QYI7 A0A154P018 A0A151I5L2 A0A2L2YAK7 A0A3S3RXK0 A0A1B0CQX2 A0A195CKY1 A0A2L2YAN4 E2AQ20 A0NBH3 A0A182VH83 A0A1S4FBV4 A0A182I3F8 A0A182KRH9 A0A182X3L2 A0A182JXT7 A0A182MUQ9 A0A182PHA0 A0A182RQU7 A0A182NH56 E0VWC3 A0A2T7NHV8 A0A182WJ39 A0A182TYE5 A0A182ITR8 A0A084WNG9 A0A0B6ZKM6 A0A1J1IVN0 A0A182GRD0 A0A1J1IV99 A0A1J1ITR1 V4A3Q5 A0A2S2P476 A0A0J7KT07 A0A182XZ10 T1K617 A0A2H8TVI2 A0A131XRB8 A0A1E1XGW7 A0A210PE59 A0A0P4WFY5 A0A2M4A0A2 A0A2M3ZZY8 A0A2M4BF85 A0A2M4A006 L7LVN6 A0A2M4BE04 W5JKV9 A0A3N0ZAF8 A0A131XGZ5 A0A2M4BD30 A0A0L8IB18 L7LVP1 A0A2M3YZ65 A0A336LCW4 A0A182QGD4 A0A3Q2Z4E2 W5MJ61 W5LEF5 H2T7H6 C3YFB3 A0A3S1AZB3 A0A3Q1J470 A0A3B3ZZ38 A0A336MZK3 A0A0P7VGX5
D6WPF0 A0A212EJZ6 A0A1B6E6P4 A0A1Y1L8E1 V9IK44 A0A0M8ZQ75 A0A1Z5LFT8 A0A0T6BAV7 A0A293LCR4 A0A1L8DSF2 A0A1L8DSA8 A0A1Z5L2N8 A0A2R5LBR3 A0A131YXB9 A0A224YUB1 A0A151X5K0 A0A0L7R0D4 A0A2I9LNQ8 A0A147BG56 A0A026WKE0 A0A023FD51 A0A195DRL1 E2BPI8 A0A195EVS6 F4X271 A0A158NAK6 A0A0C9QYI7 A0A154P018 A0A151I5L2 A0A2L2YAK7 A0A3S3RXK0 A0A1B0CQX2 A0A195CKY1 A0A2L2YAN4 E2AQ20 A0NBH3 A0A182VH83 A0A1S4FBV4 A0A182I3F8 A0A182KRH9 A0A182X3L2 A0A182JXT7 A0A182MUQ9 A0A182PHA0 A0A182RQU7 A0A182NH56 E0VWC3 A0A2T7NHV8 A0A182WJ39 A0A182TYE5 A0A182ITR8 A0A084WNG9 A0A0B6ZKM6 A0A1J1IVN0 A0A182GRD0 A0A1J1IV99 A0A1J1ITR1 V4A3Q5 A0A2S2P476 A0A0J7KT07 A0A182XZ10 T1K617 A0A2H8TVI2 A0A131XRB8 A0A1E1XGW7 A0A210PE59 A0A0P4WFY5 A0A2M4A0A2 A0A2M3ZZY8 A0A2M4BF85 A0A2M4A006 L7LVN6 A0A2M4BE04 W5JKV9 A0A3N0ZAF8 A0A131XGZ5 A0A2M4BD30 A0A0L8IB18 L7LVP1 A0A2M3YZ65 A0A336LCW4 A0A182QGD4 A0A3Q2Z4E2 W5MJ61 W5LEF5 H2T7H6 C3YFB3 A0A3S1AZB3 A0A3Q1J470 A0A3B3ZZ38 A0A336MZK3 A0A0P7VGX5
Pubmed
28756777
26354079
24845553
18362917
19820115
22118469
+ More
28004739 28528879 26830274 28797301 29248469 29652888 24508170 30249741 20798317 21719571 21347285 26561354 12364791 14747013 17210077 20966253 20566863 24438588 26483478 23254933 25244985 28503490 28812685 25576852 20920257 23761445 28049606 25329095 21551351 18563158 25463417
28004739 28528879 26830274 28797301 29248469 29652888 24508170 30249741 20798317 21719571 21347285 26561354 12364791 14747013 17210077 20966253 20566863 24438588 26483478 23254933 25244985 28503490 28812685 25576852 20920257 23761445 28049606 25329095 21551351 18563158 25463417
EMBL
KZ149915
PZC77976.1
KQ459598
KPI94499.1
NEVH01025130
PNF15964.1
+ More
GECZ01005489 JAS64280.1 GECU01018121 JAS89585.1 KK852779 KDR16690.1 KQ971354 EFA06236.2 AGBW02014361 OWR41801.1 GEDC01012053 GEDC01003719 JAS25245.1 JAS33579.1 GEZM01062288 JAV69864.1 JR051098 AEY61430.1 KQ436240 KOX67339.1 GFJQ02000714 JAW06256.1 LJIG01002491 KRT84449.1 GFWV01000888 MAA25618.1 GFDF01004715 JAV09369.1 GFDF01004716 JAV09368.1 GFJQ02005415 JAW01555.1 GGLE01002783 MBY06909.1 GEDV01005986 JAP82571.1 GFPF01006264 MAA17410.1 KQ982519 KYQ55559.1 KQ414670 KOC64298.1 GFWZ01000035 MBW20025.1 GEGO01006129 JAR89275.1 KK107167 QOIP01000010 EZA56448.1 RLU17981.1 GBBK01005217 JAC19265.1 KQ980612 KYN15139.1 GL449633 EFN82392.1 KQ981954 KYN32246.1 GL888565 EGI59487.1 ADTU01010349 ADTU01010350 GBYB01008839 JAG78606.1 KQ434781 KZC04470.1 KQ976413 KYM90384.1 IAAA01010159 IAAA01010160 IAAA01010161 LAA05199.1 NCKU01003566 NCKU01003419 RWS07281.1 RWS07546.1 AJWK01024174 AJWK01024175 AJWK01024176 AJWK01024177 AJWK01024178 AJWK01024179 AJWK01024180 AJWK01024181 AJWK01024182 AJWK01024183 KQ977642 KYN01112.1 IAAA01010162 LAA05204.1 GL441651 EFN64463.1 AAAB01008807 EAU77606.2 APCN01000208 AXCM01000681 DS235818 EEB17679.1 PZQS01000012 PVD20759.1 ATLV01024602 KE525352 KFB51763.1 HACG01022201 HACG01022203 HACG01022204 CEK69066.1 CEK69068.1 CEK69069.1 CVRI01000059 CRL03636.1 JXUM01082259 JXUM01082260 KQ563300 KXJ74101.1 CRL03634.1 CRL03635.1 KB202823 ESO87841.1 GGMR01011650 MBY24269.1 LBMM01003442 KMQ93532.1 CAEY01001591 GFXV01006470 MBW18275.1 GEFM01005912 JAP69884.1 GFAC01000907 JAT98281.1 NEDP02076750 OWF34775.1 GDRN01025825 JAI67701.1 GGFK01000747 MBW34068.1 GGFK01000721 MBW34042.1 GGFJ01002575 MBW51716.1 GGFK01000760 MBW34081.1 GACK01009164 JAA55870.1 GGFJ01002050 MBW51191.1 ADMH02001258 ETN63394.1 RJVU01000976 ROL55363.1 GEFH01002929 JAP65652.1 GGFJ01001776 MBW50917.1 KQ416096 KOF98686.1 GACK01009159 JAA55875.1 GGFM01000813 MBW21564.1 UFQS01001660 UFQT01001660 SSX11774.1 SSX31339.1 AXCN02000876 AHAT01020910 AHAT01020911 GG666509 EEN60876.1 RQTK01000693 RUS76010.1 SSX11773.1 SSX31338.1 JARO02001669 KPP74766.1
GECZ01005489 JAS64280.1 GECU01018121 JAS89585.1 KK852779 KDR16690.1 KQ971354 EFA06236.2 AGBW02014361 OWR41801.1 GEDC01012053 GEDC01003719 JAS25245.1 JAS33579.1 GEZM01062288 JAV69864.1 JR051098 AEY61430.1 KQ436240 KOX67339.1 GFJQ02000714 JAW06256.1 LJIG01002491 KRT84449.1 GFWV01000888 MAA25618.1 GFDF01004715 JAV09369.1 GFDF01004716 JAV09368.1 GFJQ02005415 JAW01555.1 GGLE01002783 MBY06909.1 GEDV01005986 JAP82571.1 GFPF01006264 MAA17410.1 KQ982519 KYQ55559.1 KQ414670 KOC64298.1 GFWZ01000035 MBW20025.1 GEGO01006129 JAR89275.1 KK107167 QOIP01000010 EZA56448.1 RLU17981.1 GBBK01005217 JAC19265.1 KQ980612 KYN15139.1 GL449633 EFN82392.1 KQ981954 KYN32246.1 GL888565 EGI59487.1 ADTU01010349 ADTU01010350 GBYB01008839 JAG78606.1 KQ434781 KZC04470.1 KQ976413 KYM90384.1 IAAA01010159 IAAA01010160 IAAA01010161 LAA05199.1 NCKU01003566 NCKU01003419 RWS07281.1 RWS07546.1 AJWK01024174 AJWK01024175 AJWK01024176 AJWK01024177 AJWK01024178 AJWK01024179 AJWK01024180 AJWK01024181 AJWK01024182 AJWK01024183 KQ977642 KYN01112.1 IAAA01010162 LAA05204.1 GL441651 EFN64463.1 AAAB01008807 EAU77606.2 APCN01000208 AXCM01000681 DS235818 EEB17679.1 PZQS01000012 PVD20759.1 ATLV01024602 KE525352 KFB51763.1 HACG01022201 HACG01022203 HACG01022204 CEK69066.1 CEK69068.1 CEK69069.1 CVRI01000059 CRL03636.1 JXUM01082259 JXUM01082260 KQ563300 KXJ74101.1 CRL03634.1 CRL03635.1 KB202823 ESO87841.1 GGMR01011650 MBY24269.1 LBMM01003442 KMQ93532.1 CAEY01001591 GFXV01006470 MBW18275.1 GEFM01005912 JAP69884.1 GFAC01000907 JAT98281.1 NEDP02076750 OWF34775.1 GDRN01025825 JAI67701.1 GGFK01000747 MBW34068.1 GGFK01000721 MBW34042.1 GGFJ01002575 MBW51716.1 GGFK01000760 MBW34081.1 GACK01009164 JAA55870.1 GGFJ01002050 MBW51191.1 ADMH02001258 ETN63394.1 RJVU01000976 ROL55363.1 GEFH01002929 JAP65652.1 GGFJ01001776 MBW50917.1 KQ416096 KOF98686.1 GACK01009159 JAA55875.1 GGFM01000813 MBW21564.1 UFQS01001660 UFQT01001660 SSX11774.1 SSX31339.1 AXCN02000876 AHAT01020910 AHAT01020911 GG666509 EEN60876.1 RQTK01000693 RUS76010.1 SSX11773.1 SSX31338.1 JARO02001669 KPP74766.1
Proteomes
UP000053268
UP000235965
UP000027135
UP000007266
UP000007151
UP000053105
+ More
UP000075809 UP000053825 UP000053097 UP000279307 UP000078492 UP000008237 UP000078541 UP000007755 UP000005205 UP000076502 UP000078540 UP000285301 UP000092461 UP000078542 UP000000311 UP000007062 UP000075903 UP000075840 UP000075882 UP000076407 UP000075881 UP000075883 UP000075885 UP000075900 UP000075884 UP000009046 UP000245119 UP000075920 UP000075902 UP000075880 UP000030765 UP000183832 UP000069940 UP000249989 UP000030746 UP000036403 UP000076408 UP000015104 UP000242188 UP000000673 UP000053454 UP000075886 UP000264820 UP000018468 UP000018467 UP000005226 UP000001554 UP000271974 UP000265040 UP000261520 UP000034805
UP000075809 UP000053825 UP000053097 UP000279307 UP000078492 UP000008237 UP000078541 UP000007755 UP000005205 UP000076502 UP000078540 UP000285301 UP000092461 UP000078542 UP000000311 UP000007062 UP000075903 UP000075840 UP000075882 UP000076407 UP000075881 UP000075883 UP000075885 UP000075900 UP000075884 UP000009046 UP000245119 UP000075920 UP000075902 UP000075880 UP000030765 UP000183832 UP000069940 UP000249989 UP000030746 UP000036403 UP000076408 UP000015104 UP000242188 UP000000673 UP000053454 UP000075886 UP000264820 UP000018468 UP000018467 UP000005226 UP000001554 UP000271974 UP000265040 UP000261520 UP000034805
Pfam
Interpro
IPR001841
Znf_RING
+ More
IPR013083 Znf_RING/FYVE/PHD
IPR003892 CUE
IPR040675 AMFR_Ube2g2-bd
IPR036462 Fumarylacetoacetase_N_sf
IPR005959 Fumarylacetoacetase
IPR015377 Fumarylacetoacetase_N
IPR036663 Fumarylacetoacetase_C_sf
IPR011234 Fumarylacetoacetase-like_C
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR002110 Ankyrin_rpt
IPR001507 ZP_dom
IPR013083 Znf_RING/FYVE/PHD
IPR003892 CUE
IPR040675 AMFR_Ube2g2-bd
IPR036462 Fumarylacetoacetase_N_sf
IPR005959 Fumarylacetoacetase
IPR015377 Fumarylacetoacetase_N
IPR036663 Fumarylacetoacetase_C_sf
IPR011234 Fumarylacetoacetase-like_C
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR002110 Ankyrin_rpt
IPR001507 ZP_dom
Gene 3D
CDD
ProteinModelPortal
A0A2W1BZ63
A0A194PP26
A0A2J7PI11
A0A1B6GPE6
A0A1B6IRM5
A0A067RD56
+ More
D6WPF0 A0A212EJZ6 A0A1B6E6P4 A0A1Y1L8E1 V9IK44 A0A0M8ZQ75 A0A1Z5LFT8 A0A0T6BAV7 A0A293LCR4 A0A1L8DSF2 A0A1L8DSA8 A0A1Z5L2N8 A0A2R5LBR3 A0A131YXB9 A0A224YUB1 A0A151X5K0 A0A0L7R0D4 A0A2I9LNQ8 A0A147BG56 A0A026WKE0 A0A023FD51 A0A195DRL1 E2BPI8 A0A195EVS6 F4X271 A0A158NAK6 A0A0C9QYI7 A0A154P018 A0A151I5L2 A0A2L2YAK7 A0A3S3RXK0 A0A1B0CQX2 A0A195CKY1 A0A2L2YAN4 E2AQ20 A0NBH3 A0A182VH83 A0A1S4FBV4 A0A182I3F8 A0A182KRH9 A0A182X3L2 A0A182JXT7 A0A182MUQ9 A0A182PHA0 A0A182RQU7 A0A182NH56 E0VWC3 A0A2T7NHV8 A0A182WJ39 A0A182TYE5 A0A182ITR8 A0A084WNG9 A0A0B6ZKM6 A0A1J1IVN0 A0A182GRD0 A0A1J1IV99 A0A1J1ITR1 V4A3Q5 A0A2S2P476 A0A0J7KT07 A0A182XZ10 T1K617 A0A2H8TVI2 A0A131XRB8 A0A1E1XGW7 A0A210PE59 A0A0P4WFY5 A0A2M4A0A2 A0A2M3ZZY8 A0A2M4BF85 A0A2M4A006 L7LVN6 A0A2M4BE04 W5JKV9 A0A3N0ZAF8 A0A131XGZ5 A0A2M4BD30 A0A0L8IB18 L7LVP1 A0A2M3YZ65 A0A336LCW4 A0A182QGD4 A0A3Q2Z4E2 W5MJ61 W5LEF5 H2T7H6 C3YFB3 A0A3S1AZB3 A0A3Q1J470 A0A3B3ZZ38 A0A336MZK3 A0A0P7VGX5
D6WPF0 A0A212EJZ6 A0A1B6E6P4 A0A1Y1L8E1 V9IK44 A0A0M8ZQ75 A0A1Z5LFT8 A0A0T6BAV7 A0A293LCR4 A0A1L8DSF2 A0A1L8DSA8 A0A1Z5L2N8 A0A2R5LBR3 A0A131YXB9 A0A224YUB1 A0A151X5K0 A0A0L7R0D4 A0A2I9LNQ8 A0A147BG56 A0A026WKE0 A0A023FD51 A0A195DRL1 E2BPI8 A0A195EVS6 F4X271 A0A158NAK6 A0A0C9QYI7 A0A154P018 A0A151I5L2 A0A2L2YAK7 A0A3S3RXK0 A0A1B0CQX2 A0A195CKY1 A0A2L2YAN4 E2AQ20 A0NBH3 A0A182VH83 A0A1S4FBV4 A0A182I3F8 A0A182KRH9 A0A182X3L2 A0A182JXT7 A0A182MUQ9 A0A182PHA0 A0A182RQU7 A0A182NH56 E0VWC3 A0A2T7NHV8 A0A182WJ39 A0A182TYE5 A0A182ITR8 A0A084WNG9 A0A0B6ZKM6 A0A1J1IVN0 A0A182GRD0 A0A1J1IV99 A0A1J1ITR1 V4A3Q5 A0A2S2P476 A0A0J7KT07 A0A182XZ10 T1K617 A0A2H8TVI2 A0A131XRB8 A0A1E1XGW7 A0A210PE59 A0A0P4WFY5 A0A2M4A0A2 A0A2M3ZZY8 A0A2M4BF85 A0A2M4A006 L7LVN6 A0A2M4BE04 W5JKV9 A0A3N0ZAF8 A0A131XGZ5 A0A2M4BD30 A0A0L8IB18 L7LVP1 A0A2M3YZ65 A0A336LCW4 A0A182QGD4 A0A3Q2Z4E2 W5MJ61 W5LEF5 H2T7H6 C3YFB3 A0A3S1AZB3 A0A3Q1J470 A0A3B3ZZ38 A0A336MZK3 A0A0P7VGX5
PDB
2LXH
E-value=1.13857e-18,
Score=231
Ontologies
GO
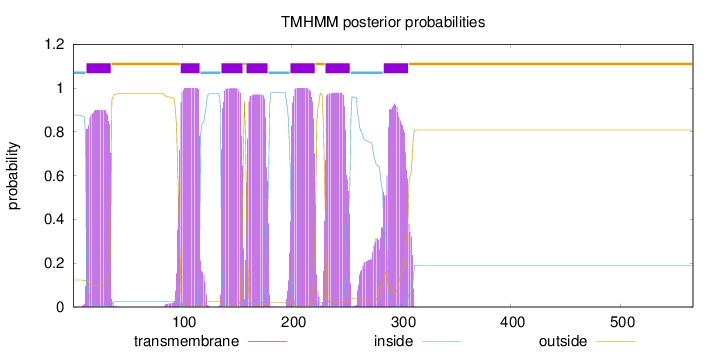
Topology
Length:
566
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
147.24887
Exp number, first 60 AAs:
20.21114
Total prob of N-in:
0.87635
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 98
TMhelix
99 - 116
inside
117 - 135
TMhelix
136 - 155
outside
156 - 158
TMhelix
159 - 178
inside
179 - 198
TMhelix
199 - 221
outside
222 - 230
TMhelix
231 - 253
inside
254 - 283
TMhelix
284 - 306
outside
307 - 566
Population Genetic Test Statistics
Pi
1.360594
Theta
4.176993
Tajima's D
-2.172887
CLR
17.075237
CSRT
0.00434978251087446
Interpretation
Possibly Positive selection
