Pre Gene Modal
BGIBMGA000188
Annotation
hypothetical_protein_KGM_02087_[Danaus_plexippus]
Full name
Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit DAD1
Alternative Name
Defender against apoptotic cell death 1
Defender against cell death 1
Defender against cell death 1
Location in the cell
PlasmaMembrane Reliability : 3.837
Sequence
CDS
ATGTCTTCACTTACTGCTGTAATTCCAAAATTGTATCAAGAATACACAACGAAAACTCCTAAGAAGCTTAAAATAATTGATGCATACCTTTTCTATATTTTCCTTACTGCGGTCATACAATTCGGCTACTGTTGCCTTGTCGGCACTTTCCCATTTAATTCTTTCTTAAGTGGTTTTATATCTACAGTAAGCTGCTTTGTTCTTGGAGTGTGCCTTCGGCTACAAGTTAATCCAGAAAACAAGAATGAGTTCCAAGGGCTGAGTGCAGAACGTGGCTTTGCAGATTTCATATTTGCACATCTGGTGTTGCATATTGTTGTTATCAATTTCATAGGATAA
Protein
MSSLTAVIPKLYQEYTTKTPKKLKIIDAYLFYIFLTAVIQFGYCCLVGTFPFNSFLSGFISTVSCFVLGVCLRLQVNPENKNEFQGLSAERGFADFIFAHLVLHIVVINFIG
Summary
Description
Subunit of the oligosaccharyl transferase (OST) complex that catalyzes the initial transfer of a defined glycan (Glc(3)Man(9)GlcNAc(2) in eukaryotes) from the lipid carrier dolichol-pyrophosphate to an asparagine residue within an Asn-X-Ser/Thr consensus motif in nascent polypeptide chains, the first step in protein N-glycosylation. N-glycosylation occurs cotranslationally and the complex associates with the Sec61 complex at the channel-forming translocon complex that mediates protein translocation across the endoplasmic reticulum (ER). All subunits are required for a maximal enzyme activity.
Subunit of the oligosaccharyl transferase (OST) complex that catalyzes the initial transfer of a defined glycan (Glc(3)Man(9)GlcNAc(2) in eukaryotes) from the lipid carrier dolichol-pyrophosphate to an asparagine residue within an Asn-X-Ser/Thr consensus motif in nascent polypeptide chains, the first step in protein N-glycosylation. N-glycosylation occurs cotranslationally and the complex associates with the Sec61 complex at the channel-forming translocon complex that mediates protein translocation across the endoplasmic reticulum (ER). All subunits are required for a maximal enzyme activity (By similarity). Probably as part of the N-glycosylation pathway, plays a role in the regulation of tissue growth and apoptosis (PubMed:27693235).
Subunit of the oligosaccharyl transferase (OST) complex that catalyzes the initial transfer of a defined glycan (Glc(3)Man(9)GlcNAc(2) in eukaryotes) from the lipid carrier dolichol-pyrophosphate to an asparagine residue within an Asn-X-Ser/Thr consensus motif in nascent polypeptide chains, the first step in protein N-glycosylation. N-glycosylation occurs cotranslationally and the complex associates with the Sec61 complex at the channel-forming translocon complex that mediates protein translocation across the endoplasmic reticulum (ER). All subunits are required for a maximal enzyme activity (By similarity). Probably as part of the N-glycosylation pathway, plays a role in the regulation of tissue growth and apoptosis (PubMed:27693235).
Subunit
Component of the oligosaccharyltransferase (OST) complex.
Similarity
Belongs to the DAD/OST2 family.
Keywords
Apoptosis
Complete proteome
Endoplasmic reticulum
Membrane
Reference proteome
Transmembrane
Transmembrane helix
Feature
chain Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit DAD1
Uniprot
H9ISG1
A0A212EJW2
I4DJZ7
A0A2A4IXI6
A0A2W1BXL9
A0A2H1W3K3
+ More
A0A3S2NPG0 S4NMX7 A0A0L7KV66 A0A0M8ZMJ2 A0A2A3E0B1 A0A088AW39 A0A1W4WN38 A0A154NY13 N6U0L6 A0A0N9QRN3 D6WP97 A0A067RD69 E2BPI7 A0A182R0G6 R4FPD9 A0A1B6IMT2 A0A182JLW4 A0A1B6EZC2 A0A195DQF9 A9YES0 A0A1B0CZR7 A0A182G077 W5JU24 A0A2M4C420 T1DHB4 A0A2M4A7Y5 A0A151I5T5 A0A158NAK7 A0A151X5K6 A0A182RRJ8 A0A182NUV7 A0A2M3Z2F3 A0A0C9R3A3 A0A182UKI3 A0A182P3B1 A0A182Y5J7 Q6TEC7 A0A182UPD2 A0A182X158 Q7Q4M9 A0A182IBW7 A0A172ZPW3 F4X272 L0H0N4 A0A195CM13 A0A2S2P6S6 A0A182M0H0 A0A182WHH6 U5EM12 A0A195EWH7 E0VWC4 A0A0L7R0A5 A0A182KAZ9 A0A2M4A7V7 A0A2H8TSN1 A0A0K8TQP8 E2AQ19 A0A026WMW4 A0A224XU03 A0A069DNT9 A0A1Q3FFA2 A0A1L8DWF0 A0A1C9CWI1 A0A0U2KE66 B0W8T7 A0A023ED79 J9JJ86 Q1HRQ3 A0A3S1BAJ2 A0A3S3P7U0 A0A0A9XSX2 A9Q6B7 B4NWY0 B4HYK4 B3N759 B4Q6S5 Q9VLM5 A0A336LXA9 A0A1J1IKT7 A0A3R7P460 A0A0A6ZDL6 A0A1S3CYY7 A0A1W4VX07 C4WW96 T1DJ44 A0A0C9R1G6 B4N0Y4 B3MP03 A0A0K8UNV5 A0A182GMW4 A0A146LR50 V3ZZ05 A0A2T7NHV4 A0A0M4ERN8
A0A3S2NPG0 S4NMX7 A0A0L7KV66 A0A0M8ZMJ2 A0A2A3E0B1 A0A088AW39 A0A1W4WN38 A0A154NY13 N6U0L6 A0A0N9QRN3 D6WP97 A0A067RD69 E2BPI7 A0A182R0G6 R4FPD9 A0A1B6IMT2 A0A182JLW4 A0A1B6EZC2 A0A195DQF9 A9YES0 A0A1B0CZR7 A0A182G077 W5JU24 A0A2M4C420 T1DHB4 A0A2M4A7Y5 A0A151I5T5 A0A158NAK7 A0A151X5K6 A0A182RRJ8 A0A182NUV7 A0A2M3Z2F3 A0A0C9R3A3 A0A182UKI3 A0A182P3B1 A0A182Y5J7 Q6TEC7 A0A182UPD2 A0A182X158 Q7Q4M9 A0A182IBW7 A0A172ZPW3 F4X272 L0H0N4 A0A195CM13 A0A2S2P6S6 A0A182M0H0 A0A182WHH6 U5EM12 A0A195EWH7 E0VWC4 A0A0L7R0A5 A0A182KAZ9 A0A2M4A7V7 A0A2H8TSN1 A0A0K8TQP8 E2AQ19 A0A026WMW4 A0A224XU03 A0A069DNT9 A0A1Q3FFA2 A0A1L8DWF0 A0A1C9CWI1 A0A0U2KE66 B0W8T7 A0A023ED79 J9JJ86 Q1HRQ3 A0A3S1BAJ2 A0A3S3P7U0 A0A0A9XSX2 A9Q6B7 B4NWY0 B4HYK4 B3N759 B4Q6S5 Q9VLM5 A0A336LXA9 A0A1J1IKT7 A0A3R7P460 A0A0A6ZDL6 A0A1S3CYY7 A0A1W4VX07 C4WW96 T1DJ44 A0A0C9R1G6 B4N0Y4 B3MP03 A0A0K8UNV5 A0A182GMW4 A0A146LR50 V3ZZ05 A0A2T7NHV4 A0A0M4ERN8
Pubmed
19121390
22118469
22651552
26354079
28756777
23622113
+ More
26227816 23537049 26428861 18362917 19820115 24845553 20798317 18249019 20920257 23761445 21347285 25244985 12364791 14747013 17210077 21719571 20566863 26369729 24508170 30249741 26334808 27582290 26621068 24945155 17204158 17510324 25401762 18083552 17994087 17550304 22936249 10731132 12537572 12537569 27693235 24330624 26483478 26823975 23254933
26227816 23537049 26428861 18362917 19820115 24845553 20798317 18249019 20920257 23761445 21347285 25244985 12364791 14747013 17210077 21719571 20566863 26369729 24508170 30249741 26334808 27582290 26621068 24945155 17204158 17510324 25401762 18083552 17994087 17550304 22936249 10731132 12537572 12537569 27693235 24330624 26483478 26823975 23254933
EMBL
BABH01040705
AGBW02014361
OWR41802.1
AK401615
KQ459598
BAM18237.1
+ More
KPI94500.1 NWSH01005842 PCG63902.1 KZ149915 PZC77977.1 ODYU01006118 SOQ47679.1 RSAL01000017 RVE52961.1 GAIX01014186 JAA78374.1 JTDY01005418 KOB67025.1 KQ436240 KOX67340.1 KZ288490 PBC25205.1 KQ434781 KZC04471.1 APGK01054200 KB741247 KB631740 ENN72072.1 ERL85692.1 KP890834 KP890835 ALH18097.1 KQ971354 EFA07387.1 KK852779 KDR16705.1 GL449633 EFN82391.1 AXCN02000747 GAHY01000929 JAA76581.1 GECU01023909 GECU01019487 JAS83797.1 JAS88219.1 GECZ01026495 JAS43274.1 KQ980612 KYN15140.1 EU188857 ABY21317.1 AJVK01009695 ADMH02000447 ETN66435.1 GGFJ01010921 MBW60062.1 GAMD01002441 JAA99149.1 GGFK01003521 MBW36842.1 KQ976413 KYM90383.1 ADTU01010351 KQ982519 KYQ55560.1 GGFM01001940 MBW22691.1 GBYB01001271 JAG71038.1 AY423354 AAQ94040.1 AAAB01008964 EAA12890.2 APCN01000752 KU351657 ANF99571.1 GL888565 EGI59488.1 JX966249 AGA92566.1 KQ977642 KYN01114.1 GGMR01012538 MBY25157.1 AXCM01001953 GANO01001197 JAB58674.1 KQ981954 KYN32247.1 DS235818 EEB17680.1 KQ414670 KOC64299.1 GGFK01003555 MBW36876.1 GFXV01005392 MBW17197.1 GDAI01001378 JAI16225.1 GL441651 EFN64462.1 KK107167 QOIP01000010 EZA56449.1 RLU17976.1 GFTR01001802 JAW14624.1 GBGD01003563 JAC85326.1 GFDL01008810 JAV26235.1 GFDF01003335 JAV10749.1 KX214530 AON76443.1 KT755280 ALS05114.1 DS231860 EDS39316.1 GAPW01006248 GAPW01006247 GAPW01006246 GAPW01006245 JAC07352.1 ABLF02036609 ABLF02036612 DQ440041 CH477236 ABF18074.1 EAT46645.1 RQTK01000693 RUS76013.1 NCKU01000818 RWS14026.1 GBHO01020545 JAG23059.1 EF581986 ABU54835.1 CM000157 EDW88510.1 CH480818 EDW52134.1 CH954177 EDV59355.1 CM000361 CM002910 EDX04234.1 KMY89067.1 AE014134 AY113429 KX531951 UFQT01000278 SSX22686.1 CVRI01000054 CRL00847.1 QCYY01001831 ROT74939.1 KC427985 AGJ03553.1 AK341934 BAH72166.1 GALA01000526 JAA94326.1 GBYB01009994 JAG79761.1 CH963920 EDW78146.1 CH902620 EDV31169.1 GDHF01024068 JAI28246.1 JXUM01075156 KQ562872 KXJ74956.1 GDHC01009393 JAQ09236.1 KB202823 ESO87840.1 PZQS01000012 PVD20760.1 CP012523 ALC39875.1 ALC39883.1 ALC40262.1
KPI94500.1 NWSH01005842 PCG63902.1 KZ149915 PZC77977.1 ODYU01006118 SOQ47679.1 RSAL01000017 RVE52961.1 GAIX01014186 JAA78374.1 JTDY01005418 KOB67025.1 KQ436240 KOX67340.1 KZ288490 PBC25205.1 KQ434781 KZC04471.1 APGK01054200 KB741247 KB631740 ENN72072.1 ERL85692.1 KP890834 KP890835 ALH18097.1 KQ971354 EFA07387.1 KK852779 KDR16705.1 GL449633 EFN82391.1 AXCN02000747 GAHY01000929 JAA76581.1 GECU01023909 GECU01019487 JAS83797.1 JAS88219.1 GECZ01026495 JAS43274.1 KQ980612 KYN15140.1 EU188857 ABY21317.1 AJVK01009695 ADMH02000447 ETN66435.1 GGFJ01010921 MBW60062.1 GAMD01002441 JAA99149.1 GGFK01003521 MBW36842.1 KQ976413 KYM90383.1 ADTU01010351 KQ982519 KYQ55560.1 GGFM01001940 MBW22691.1 GBYB01001271 JAG71038.1 AY423354 AAQ94040.1 AAAB01008964 EAA12890.2 APCN01000752 KU351657 ANF99571.1 GL888565 EGI59488.1 JX966249 AGA92566.1 KQ977642 KYN01114.1 GGMR01012538 MBY25157.1 AXCM01001953 GANO01001197 JAB58674.1 KQ981954 KYN32247.1 DS235818 EEB17680.1 KQ414670 KOC64299.1 GGFK01003555 MBW36876.1 GFXV01005392 MBW17197.1 GDAI01001378 JAI16225.1 GL441651 EFN64462.1 KK107167 QOIP01000010 EZA56449.1 RLU17976.1 GFTR01001802 JAW14624.1 GBGD01003563 JAC85326.1 GFDL01008810 JAV26235.1 GFDF01003335 JAV10749.1 KX214530 AON76443.1 KT755280 ALS05114.1 DS231860 EDS39316.1 GAPW01006248 GAPW01006247 GAPW01006246 GAPW01006245 JAC07352.1 ABLF02036609 ABLF02036612 DQ440041 CH477236 ABF18074.1 EAT46645.1 RQTK01000693 RUS76013.1 NCKU01000818 RWS14026.1 GBHO01020545 JAG23059.1 EF581986 ABU54835.1 CM000157 EDW88510.1 CH480818 EDW52134.1 CH954177 EDV59355.1 CM000361 CM002910 EDX04234.1 KMY89067.1 AE014134 AY113429 KX531951 UFQT01000278 SSX22686.1 CVRI01000054 CRL00847.1 QCYY01001831 ROT74939.1 KC427985 AGJ03553.1 AK341934 BAH72166.1 GALA01000526 JAA94326.1 GBYB01009994 JAG79761.1 CH963920 EDW78146.1 CH902620 EDV31169.1 GDHF01024068 JAI28246.1 JXUM01075156 KQ562872 KXJ74956.1 GDHC01009393 JAQ09236.1 KB202823 ESO87840.1 PZQS01000012 PVD20760.1 CP012523 ALC39875.1 ALC39883.1 ALC40262.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000218220
UP000283053
UP000037510
+ More
UP000053105 UP000242457 UP000005203 UP000192223 UP000076502 UP000019118 UP000030742 UP000007266 UP000027135 UP000008237 UP000075886 UP000075880 UP000078492 UP000092462 UP000069272 UP000000673 UP000078540 UP000005205 UP000075809 UP000075900 UP000075884 UP000075902 UP000075885 UP000076408 UP000075903 UP000076407 UP000007062 UP000075840 UP000007755 UP000078542 UP000075883 UP000075920 UP000078541 UP000009046 UP000053825 UP000075881 UP000000311 UP000053097 UP000279307 UP000002320 UP000007819 UP000008820 UP000271974 UP000285301 UP000002282 UP000001292 UP000008711 UP000000304 UP000000803 UP000183832 UP000283509 UP000079169 UP000192221 UP000007798 UP000007801 UP000069940 UP000249989 UP000030746 UP000245119 UP000092553
UP000053105 UP000242457 UP000005203 UP000192223 UP000076502 UP000019118 UP000030742 UP000007266 UP000027135 UP000008237 UP000075886 UP000075880 UP000078492 UP000092462 UP000069272 UP000000673 UP000078540 UP000005205 UP000075809 UP000075900 UP000075884 UP000075902 UP000075885 UP000076408 UP000075903 UP000076407 UP000007062 UP000075840 UP000007755 UP000078542 UP000075883 UP000075920 UP000078541 UP000009046 UP000053825 UP000075881 UP000000311 UP000053097 UP000279307 UP000002320 UP000007819 UP000008820 UP000271974 UP000285301 UP000002282 UP000001292 UP000008711 UP000000304 UP000000803 UP000183832 UP000283509 UP000079169 UP000192221 UP000007798 UP000007801 UP000069940 UP000249989 UP000030746 UP000245119 UP000092553
Pfam
PF02109 DAD
Interpro
IPR003038
DAD/Ost2
ProteinModelPortal
H9ISG1
A0A212EJW2
I4DJZ7
A0A2A4IXI6
A0A2W1BXL9
A0A2H1W3K3
+ More
A0A3S2NPG0 S4NMX7 A0A0L7KV66 A0A0M8ZMJ2 A0A2A3E0B1 A0A088AW39 A0A1W4WN38 A0A154NY13 N6U0L6 A0A0N9QRN3 D6WP97 A0A067RD69 E2BPI7 A0A182R0G6 R4FPD9 A0A1B6IMT2 A0A182JLW4 A0A1B6EZC2 A0A195DQF9 A9YES0 A0A1B0CZR7 A0A182G077 W5JU24 A0A2M4C420 T1DHB4 A0A2M4A7Y5 A0A151I5T5 A0A158NAK7 A0A151X5K6 A0A182RRJ8 A0A182NUV7 A0A2M3Z2F3 A0A0C9R3A3 A0A182UKI3 A0A182P3B1 A0A182Y5J7 Q6TEC7 A0A182UPD2 A0A182X158 Q7Q4M9 A0A182IBW7 A0A172ZPW3 F4X272 L0H0N4 A0A195CM13 A0A2S2P6S6 A0A182M0H0 A0A182WHH6 U5EM12 A0A195EWH7 E0VWC4 A0A0L7R0A5 A0A182KAZ9 A0A2M4A7V7 A0A2H8TSN1 A0A0K8TQP8 E2AQ19 A0A026WMW4 A0A224XU03 A0A069DNT9 A0A1Q3FFA2 A0A1L8DWF0 A0A1C9CWI1 A0A0U2KE66 B0W8T7 A0A023ED79 J9JJ86 Q1HRQ3 A0A3S1BAJ2 A0A3S3P7U0 A0A0A9XSX2 A9Q6B7 B4NWY0 B4HYK4 B3N759 B4Q6S5 Q9VLM5 A0A336LXA9 A0A1J1IKT7 A0A3R7P460 A0A0A6ZDL6 A0A1S3CYY7 A0A1W4VX07 C4WW96 T1DJ44 A0A0C9R1G6 B4N0Y4 B3MP03 A0A0K8UNV5 A0A182GMW4 A0A146LR50 V3ZZ05 A0A2T7NHV4 A0A0M4ERN8
A0A3S2NPG0 S4NMX7 A0A0L7KV66 A0A0M8ZMJ2 A0A2A3E0B1 A0A088AW39 A0A1W4WN38 A0A154NY13 N6U0L6 A0A0N9QRN3 D6WP97 A0A067RD69 E2BPI7 A0A182R0G6 R4FPD9 A0A1B6IMT2 A0A182JLW4 A0A1B6EZC2 A0A195DQF9 A9YES0 A0A1B0CZR7 A0A182G077 W5JU24 A0A2M4C420 T1DHB4 A0A2M4A7Y5 A0A151I5T5 A0A158NAK7 A0A151X5K6 A0A182RRJ8 A0A182NUV7 A0A2M3Z2F3 A0A0C9R3A3 A0A182UKI3 A0A182P3B1 A0A182Y5J7 Q6TEC7 A0A182UPD2 A0A182X158 Q7Q4M9 A0A182IBW7 A0A172ZPW3 F4X272 L0H0N4 A0A195CM13 A0A2S2P6S6 A0A182M0H0 A0A182WHH6 U5EM12 A0A195EWH7 E0VWC4 A0A0L7R0A5 A0A182KAZ9 A0A2M4A7V7 A0A2H8TSN1 A0A0K8TQP8 E2AQ19 A0A026WMW4 A0A224XU03 A0A069DNT9 A0A1Q3FFA2 A0A1L8DWF0 A0A1C9CWI1 A0A0U2KE66 B0W8T7 A0A023ED79 J9JJ86 Q1HRQ3 A0A3S1BAJ2 A0A3S3P7U0 A0A0A9XSX2 A9Q6B7 B4NWY0 B4HYK4 B3N759 B4Q6S5 Q9VLM5 A0A336LXA9 A0A1J1IKT7 A0A3R7P460 A0A0A6ZDL6 A0A1S3CYY7 A0A1W4VX07 C4WW96 T1DJ44 A0A0C9R1G6 B4N0Y4 B3MP03 A0A0K8UNV5 A0A182GMW4 A0A146LR50 V3ZZ05 A0A2T7NHV4 A0A0M4ERN8
PDB
6EZN
E-value=1.89621e-12,
Score=168
Ontologies
PATHWAY
GO
PANTHER
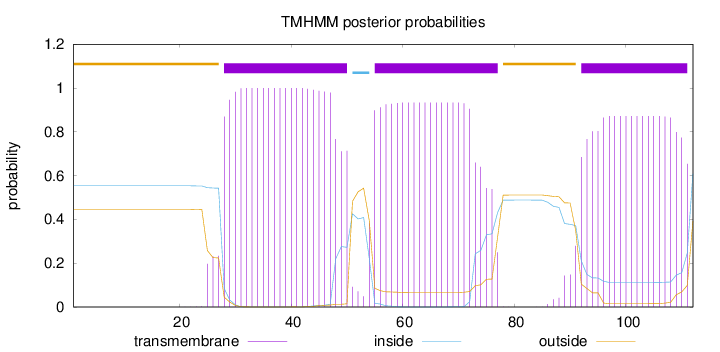
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
112
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
59.78218
Exp number, first 60 AAs:
28.71387
Total prob of N-in:
0.55403
POSSIBLE N-term signal
sequence
outside
1 - 27
TMhelix
28 - 50
inside
51 - 54
TMhelix
55 - 77
outside
78 - 91
TMhelix
92 - 111
inside
112 - 112
Population Genetic Test Statistics
Pi
17.022726
Theta
14.401598
Tajima's D
0.098156
CLR
0.210041
CSRT
0.408929553522324
Interpretation
Uncertain
